COVID-19 disease begins as a respiratory infection, in the nasal cavity, with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral replication continuing in the nasal epithelium for days and eventually spreading downwards to involve the lower respiratory tract. Scientists are therefore wondering if administering a viral inhibitor into the nose and respiratory system would work well to prevent infection or to treat early SARS-CoV-2 infection. In this case, such a drug would be valuable in treating healthcare workers and others at high risk.
The new molecules are small stable compounds that bind to the spike protein at high affinity. Since they are capable of binding to multiple domains, they allow the density of inhibitory domains to be maximal. Thus, they can be instilled intranasally, or administered by nebulization or as a dry powder aerosol, preventing the viral spike protein from binding to the host ACE2 receptor.
The study carries forwards the findings of a previous influenza study in which similar small proteins designed for tight interactions with the hemagglutinin of the flu virus was able to both prevent and treat deadly influenza in experimental rodent models.
Two Approaches to Minibinder Design
The researchers aimed to create such mini-binder molecules with a high affinity for the spike receptor-binding domain (RBD) to inhibit ACE2 binding competitively. They followed two directions: one was based on scaffolds built around the alpha-helix from the ACE2 protein that is responsible for most interactions with the RBD, incorporating it into small proteins designed to have more binding sites for the RBD and thus have increased affinity.
The second approach was to design de novo mini-binders irrespective of identified interactions with the RBD, so as to explore a much broader range of potential structures with many more modes of binding at high affinity to the RBD regions around the site of binding to the ACE2 receptor. This would theoretically prevent the mutational escape of the virus from neutralization.
Finding Promising Candidates
They came up with three designs and 105 designs by the first and second approaches, respectively. These were incorporated into long oligonucleotides, which were examined for their binding to RBD on the yeast cell surface, tagged with fluorescent dye for easy identification of mini-binder-RBD binding. All the first three and 12 of the second type of design were encoded and expressed by E. coli to measure the binding affinity.
The researchers found a binding affinity of ~2 uM and from 100nM to 2uM with one of the alpha-helix-based designs and 11 of the de novo designs, respectively. The mutagenesis-mediated refining of the first ACE2-based design produced a variant with an even higher affinity of ~1 nM, inhibiting the ACE2-RBD binding.
Further mutagenesis-based refining of the second type of mini-binders showed that 40 of the 50 compounds thus refined showed high conservation of amino acids at both the binding site and the protein core. When the most common substitutions were built up into libraries for affinity-binding screening at decreasing concentrations, the researchers found a small number of closely related sequences. When one of each sequence was selected for each design, they found that this final subset showed high-affinity binding for the RBD competitively with ACE2.
Finally, the researchers expressed these molecules LCB1-LCB8 in E. coli and measured their affinity for the RBD. They found that the binding affinity ranged from below 1 nM to 20 nM. In fact, two of them had affinity too strong for reliable assay with the technique used. These two, LCB2 and LCB3 bound at picomolar concentrations of RBD. These were found to be stable and retained original binding activity after remaining at room temperature for 2 weeks. This may obviate the requirement for a cold chain.
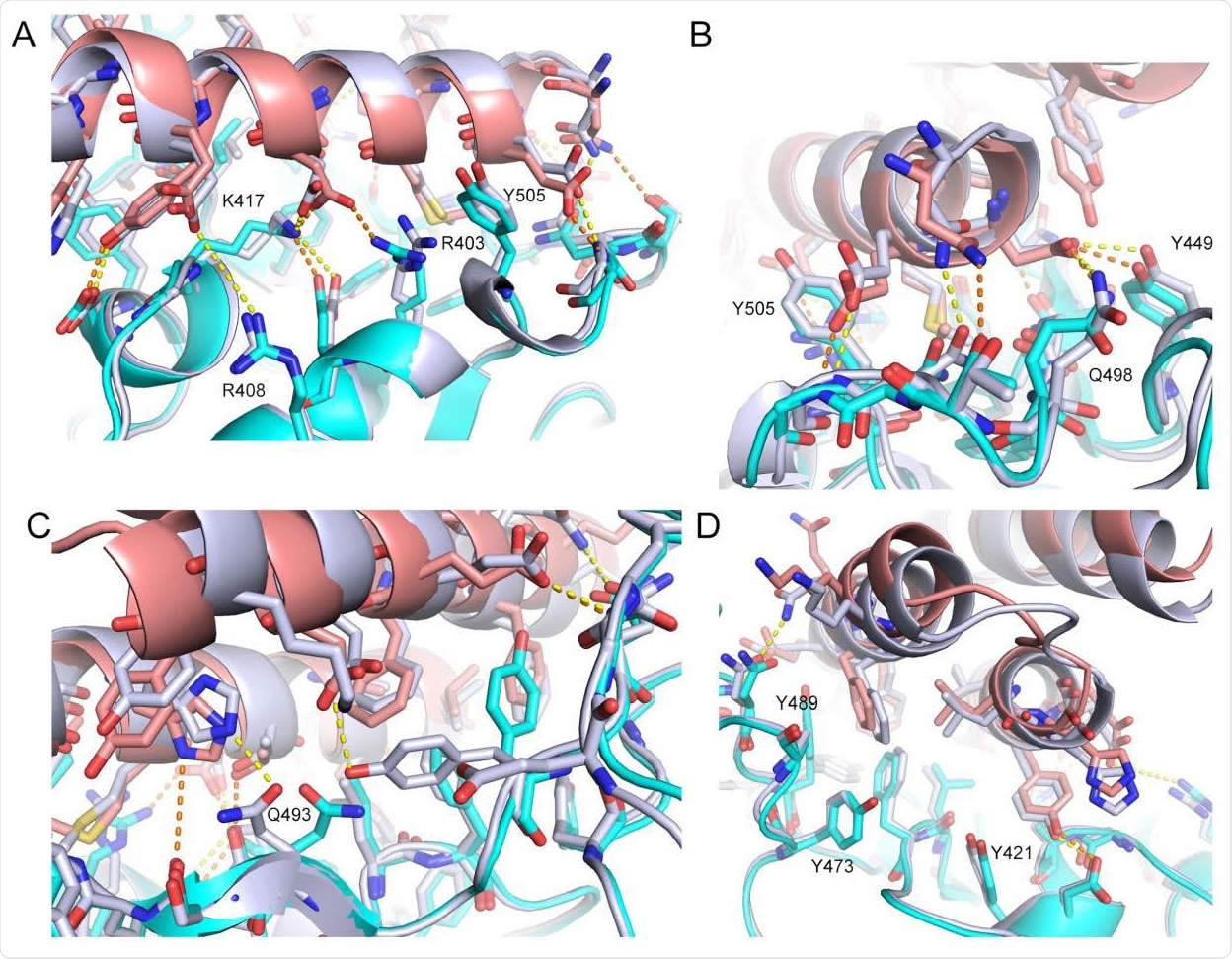
The CryoEM structure of LCB3 matches with the de novo designed model with atomic accuracy. The computational model (silver grey) of LCB3/RBD complex is overlaid with its CryoEM structure (cyan for RBD and light pink for LCB3). Interface interaction details are shown from 4 different views (A-D). Polar interactions are highlighted as dashed lines in orange for the CryoEM structure and in yellow for the design model. Representative residue indices are labeled according to the PDB 6M0J.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Structural Modeling of the Minibinder-RBD Complex
The structural evaluation showed that LCB1 and LCB3 docked to the RBD at the receptor-binding motif (RBM), which forms a small opening. These two molecules bind at opposite orientations using many electrostatic interactions, as well as taking advantage of complementary shapes in two of the alpha-helices of the mini-binders. The sites that bind to these two molecules are buried when the S protein is in the closed conformation. Only if two or more RBDs are in the ‘up state’ is it possible for the three binding sites to be recognized at the same time.
These mini-binders form both many salt bridges and hydrogen bonds with the RBM, which is responsible for the high affinities at less than nanomoles of the RBD. These two molecules have binding sites on the spike protein that overlap the binding site of ACE2, enabling competitive inhibition of virus binding to the host cell surface.
The cryoEM structures were compared with the designed models, showing close matches in the bound state and confirming the majority of polar bonds seen. Thus, the accuracy of this computational design-based method of producing high-affinity inhibitors is confirmed, with very fine variations in the form of substitutions that enhanced the affinity.
Picomolar Affinity, Neutralizing Activity
The mini-binders produced by both approaches showed potent neutralizing activity against SARS-CoV-2, but the LCB1-5 had even more potent neutralization than the first approach. The IC50 was 35 nM and 35-48 pM for the first and second approach mini-binders, respectively.
The researchers point out, “On a molar basis, these values are approximately 6-fold lower than the most potent anti-SARS-CoV-2 monoclonal antibody described to date (13); on a mass basis, because of their very small size, the designs are more potent than any of these antibodies.” In fact, each mini-binder is only a twentieth the size of a full antibody molecule, allowing them to bind to 20 times more neutralizing sites and thus conferring much higher effectiveness.
Cost-Effective and Scalable Production
Finally, the cost and ease of scalable production are much improved with these simpler molecules, which do not need to be expressed in mammal cells for proper protein folding to take place, unlike antibodies. Being small stable molecules, it should be easy to incorporate them into a nasal gel, nebulization solution, or dry powder. Moreover, they may not induce an immune reaction, as has been observed with other small de novo designed proteins, because of the small size, high stability, and good solubility which prevents dendritic cells from presenting them as antigens.
Future Directions
The speed of development of these models is striking, within 6 months. The researchers predict still faster workflows as work continues along this trajectory. This would be due to multiple reasons: more accurate methods of structure prediction, which could allow design models to be created within a day of finding the genome structure of a new pathogen.
The workflow can be decluttered to focus on identifying the optimal candidates with the best substitutions for increased affinity. This is because the cryoEM agrees well with the computational design, showing that it is relatively easy to design protein mini-binders with complementary shape and chemical sequence accurately.
The researchers predict, “With continued methods development, we believe that it will become possible to generate ultra-high-affinity, pathogen neutralizing designs within weeks of obtaining genome sequence.” This would be an invaluable asset in preparing for future pandemics caused by novel pathogens.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Baker, D. et al. (2020). De Novo Design Of Picomolar SARS-Cov-2 Miniprotein Inhibitors. bioRxiv preprint. doi: https://doi.org/10.1101/2020.08.03.234914.https://www.biorxiv.org/content/10.1101/2020.08.03.234914v1
- Peer reviewed and published scientific report.
Cao, Longxing, Inna Goreshnik, Brian Coventry, James Brett Case, Lauren Miller, Lisa Kozodoy, Rita E. Chen, et al. 2020. “De Novo Design of Picomolar SARS-CoV-2 Miniprotein Inhibitors.” Science, September. https://doi.org/10.1126/science.abd9909. https://www.science.org/doi/10.1126/science.abd9909.