A recurring concern during any viral epidemic, and especially amidst the ongoing COVID-19 pandemic, is whether we will be faced with mutations that have the propensity to alter the fundamental properties of the causative viral agent.
Already in late February, a non-synonymous mutation in the SARS-CoV-2 spike protein was noticed in viruses that were sampled from individuals in China and Europe. This mutation conferred a change from aspartic acid to glycine at position 614 of the spike protein (D614G).
Global patterns of spike 614G variant dispersal are suggestive of a selective advantage of this variant; still, the origin of 614G can be linked to early colonization events in Europe and subsequent dispersal to the rest of the world.
However, at the moment, there is no scientific consensus on the effect of the D614G mutation on SARS-CoV-2 infections, and experts are indeed skeptical that it could even produce a meaningful effect population-wise in an already highly transmissible and rapidly spreading viral illness.
Furthermore, phylodynamic estimates of reproduction numbers are rather sensitive when observed in the context of the early spread of epidemic clusters that may have involved super-spreading events.
This is why a group of researchers led by Dr. Erik M. Volz from Imperial College London decided to investigate the hypothesis for positive selection of 614G variants at the level of an individual country – the United Kingdom.
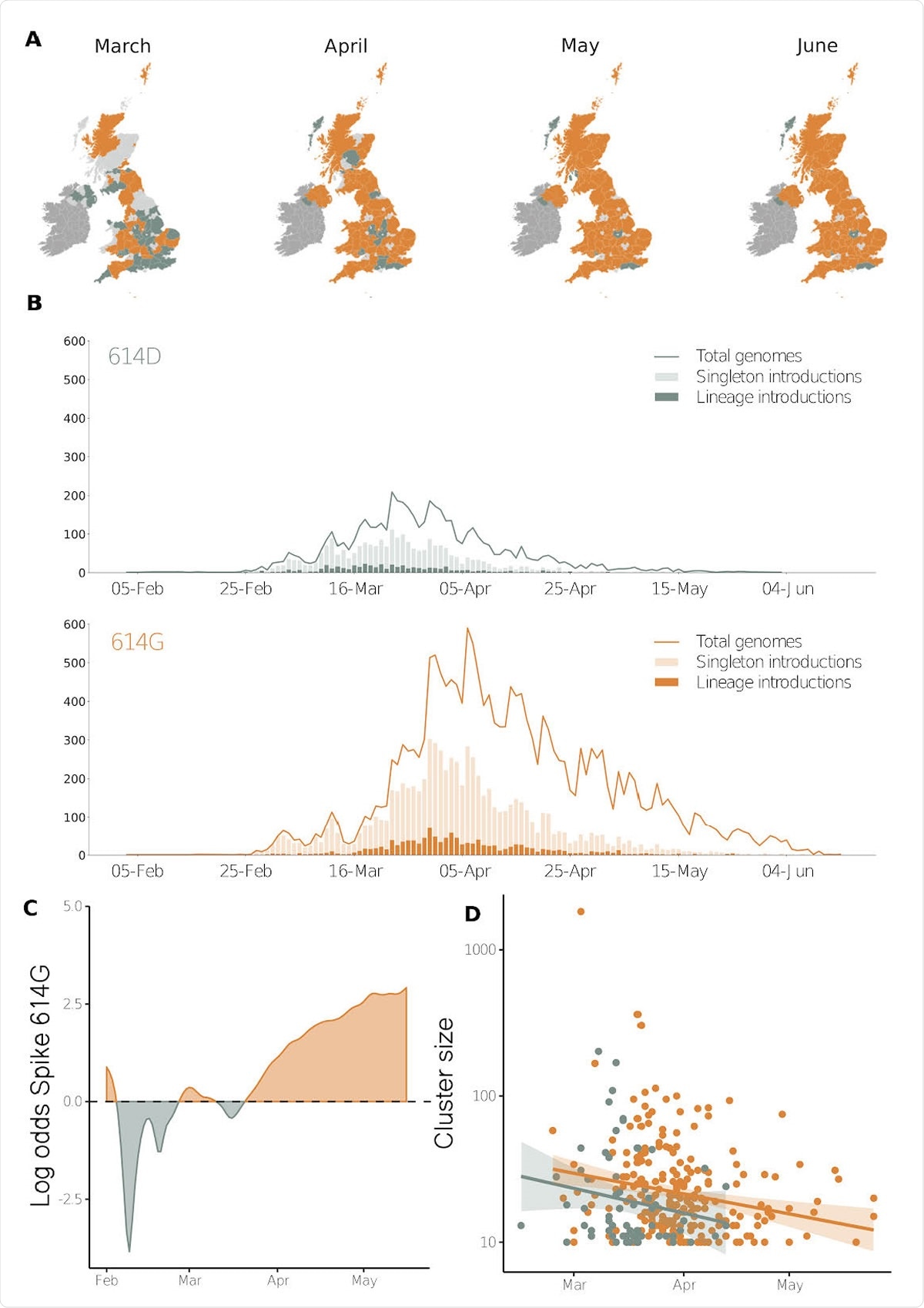
Geographic and temporal distribution of Spike 614 amino acid D or replacement G and time of lineage introductions into the United Kingdom. A) Shaded regions show the predominant residue in each region on the 15th of each month for March, April, May and June 2020, with orange indicating that 614G was more sampled and green indicating that 614D was more sampled, or that they were equally sampled. Light grey indicates that no sequences had been sampled by that point in time, and dark grey indicates the Republic of Ireland. B) The time when an introduction is first detected in the United Kingdom for Spike variants 614D and 614G. Singleton introductions are sampled exactly once and not observed subsequently, and lineage introductions gave rise to multiple sampled genomes. Solid lines show the total number of sequences collected by day of each 614 variant. C) The log odds of sampling a Spike 614G variant over time. D) The size of cluster versus time of first sample collected within a cluster.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
High-resolution dataset to examine viral changes
In their study approach, the researchers made use of the COVID-19 Genomics UK (COG-UK) SARS-CoV-2 dataset – which contains approximately half of all genomes that were sequenced globally as of July 7, 2020 – to appraise the evidence for increased transmissibility due to mutational changes in the spike protein.
This dataset has opened the door for using consistent sampling, joint laboratory, and bioinformatic approaches, as well as the collection of the same core set of metadata for all samples. Consequently, they had a sizeable, high-resolution dataset tailored to enable the examination of viral biology changes in the United Kingdom.
From there, they have estimated phylogenetic clusters within the United Kingdom by employing a maximum-parsimony reconstruction of the location of lineages within the global SARS-CoV-2 phylogeny.
Nonetheless, evaluating the epidemiological fitness of individual genetic variants during an emerging pandemic definitely presents a plethora of challenges, which the researchers took into account in their study approach.
"Overall genetic diversity of SARS-CoV-2 is low, and most methods for identifying selection will have low sensitivity given the brief time over which evolution in human hosts has been observed", caution study authors in their medRxiv paper.
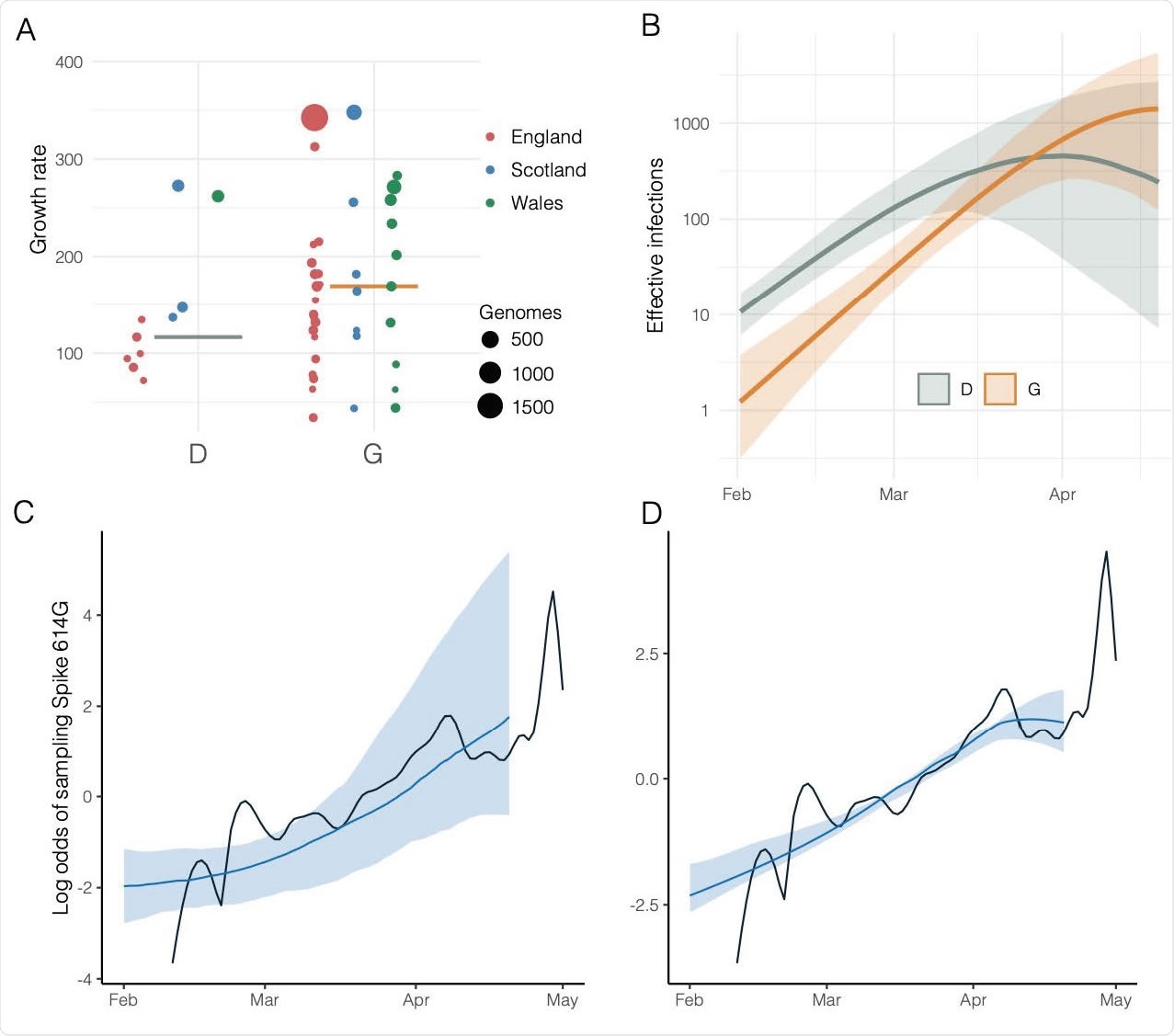
Estimated growth rates, effective infections over time, and estimated versus actual odds of sampling Spike 614G over time. A) Estimated growth rates (1/year) for 48 UK clusters at the estimated TMRCA of each cluster. Size and colour of points indicate the number of samples from each lineage and the country with more than 50% of samples. B) Estimated effective infections for a phylodynamic SEIR model fitted to 200 London sequences and 100 sequences from outside of London. Orange and green show respectively the number of 614G and 614D infections and shaded regions show 95% credible intervals. C) The estimated log odds of sampling 614G over time based on the relative size of 614G and 614D populations shown in panel (B). The shaded region shows the 95% credible interval and the black line shows the empirical log odds of sampling 614G based on a random sample of 1000 sequences. D) As panel (C) and incorporating information about observed odds of sampling 614G over time.
The effect on SARS-CoV-2 transmissibility
In a nutshell, the researchers have found that spike 614G variant clusters have been introduced in the United Kingdom later on average than 614D variant clusters, and have also grown to a larger size after adjusting for time of introduction.
Still, the phylodynamic analysis did not show a significant growth rate increase for clusters with the 614G variant. However, population genetic modeling stipulates that 614G increases in frequency relative to 614D in a manner that is consistent with a selective advantage.
In any case, there was no indication that individuals infected with the spike 614G variant had a higher risk of dying from COVID-19, albeit younger patients presented with a slightly increased odds of 614G carriage.
"Our analysis emphasizes that, while laboratory experiments can identify changes in virus biology, examining the magnitude of phenotypic effects of mutations in vitro needs to be cautiously extrapolated to predictions about transmission at a population level," say study authors.
Even though the results of this study hint that the effect on SARS-CoV-2 transmissibility caused by D614G is likely present, it is pivotal to note that the assessment of the absolute effect size is uncertain and hard to predict.
The importance for pandemic response
This study and similar research endeavors by others reveal that the transmissibility of SARS-CoV-2 can change as the pandemic unfolds. At the moment, D614G is a dominant variant – either as a primary driver of the pandemic worldwide or just a byproduct of being in the right place at the right time.
"Changes in the transmissibility of a circulating virus could have a major effect on pandemic planning and the effectiveness of the pandemic response, so it is critical that the model parameters in use for planning are based on the currently circulating virus," accentuate study authors.
Going forward, the work on drugs, vaccines, and other interventions must take into account these findings and keep in mind that reversions and other mutations (at the same or adjacent viral residues) will undoubtedly emerge in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources