New Zealand, an island country in the southwestern Pacific Ocean, is marked by its early and fierce determination to totally arrest the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the viral illness that is causing the current COVID-19 pandemic.
Genomics has played a vital role in tracing the spread and prevalence clusters of the disease, as well as the various mutations it has undergone.
Now a new study published on the preprint server medRxiv* in August 2020 reports the utility of genomics in studying the epidemiology of the virus, from research in New Zealand.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The New Zealand Outbreak – Genomic Detectives
The first SARS-CoV-2 case was reported at the end of February 2020, and New Zealand responded with a national lockdown of all services deemed non-essential. The current research focuses on 56 whole-genome sequences retrieved from as many cases in this country, combining them with epidemiologic information to arrive at an understanding of where the virus came from, over what time scale, and how it entered the country.
Between the first case and July 1, 2020, abround 1,200 cases were confirmed, 55% female, 45% male. Most cases were aged 20-29 years, and about 37% had a history of foreign travel.
The most significant number of new cases was in the Southern District Health Board, which less densely populated than many NZ cities. The highest level of lockdown was declared on March 25, just before the day on which the highest number of confirmed cases was reported. The reported incidence reached zero on May 23 and remained at that level until June 16.
On this date, new cases were found, related to overseas travel, again. However, every case from then on has been diagnosed within quarantine centers. All cases in the current study were taken from the period between the first case and the last case outside these facilities, from about 650 genomes. This represents ~56% of all confirmed cases in this period.
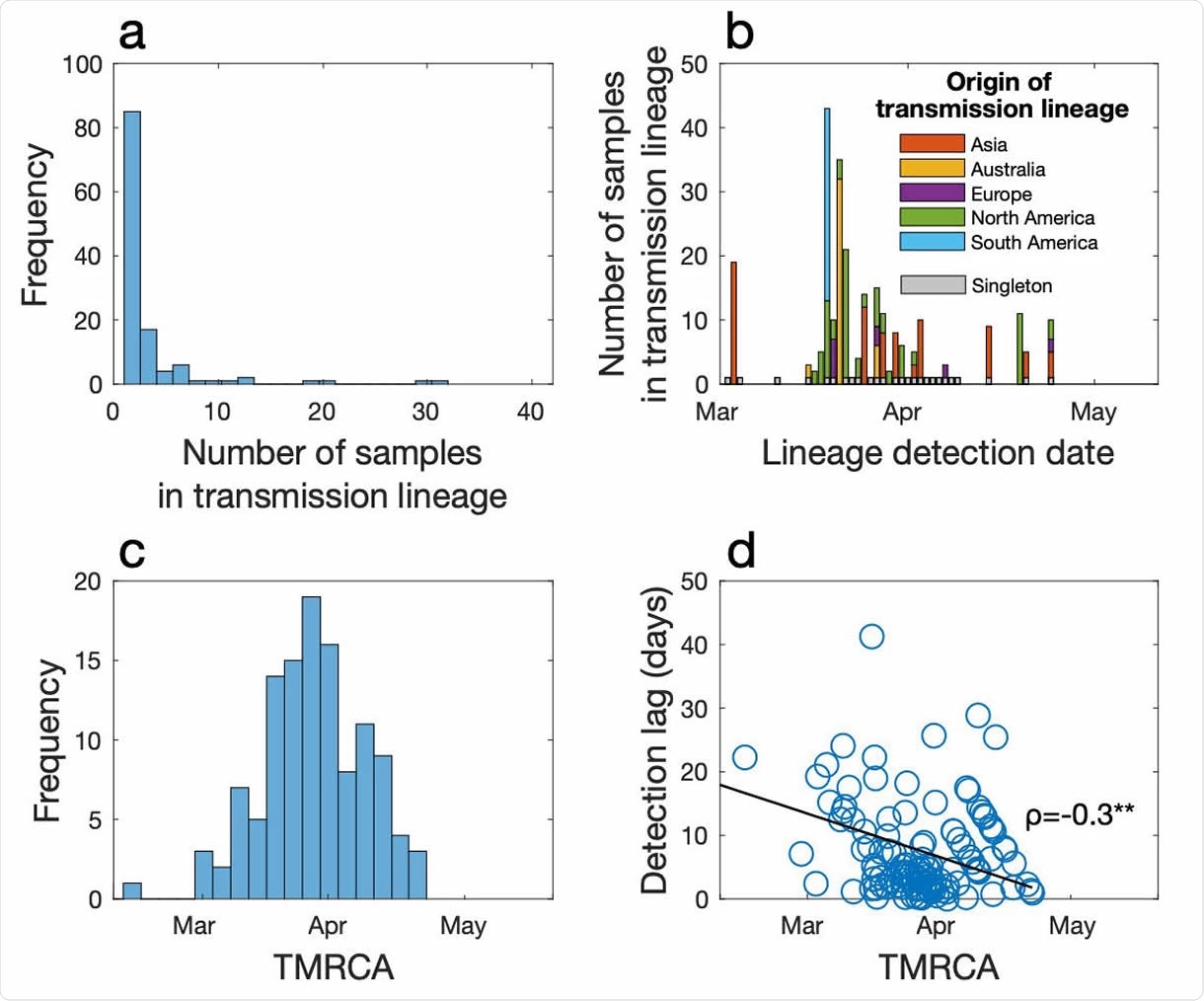
(a) Frequency of transmission lineage size. (b) The number of samples in each transmission lineage as a function of the date at which the transmission lineage was sampled, coloured by the likely origin of each lineage (inferred from epidemiological data). Importation events that led to only one additional case (singletons) are shown in grey over time. (c) Frequency of TMRCA (the time of the most recent common ancestor) of importation events over time. (d) The difference between the TMRCA and the date as which a transmission lineage was detected (i.e. detection lag) as a function of TMRCA. Spearman’s ρ indicates a significant negative relationship (p<0.01).
Viruses in NZ Represent Globally Present Strains
The viral genomes from NZ were found to represent almost every strain sequenced so far globally. The genomes had 24% aspartic acid, SD614, and 73% glycine SG614 at the 614 positions of the spike protein. This mutation has been linked to higher infectivity in cell cultures.
This increase in the frequency of glycine indicates that this variant was imported several times into NZ, and not that this strain was selected for. Moreover, the temporal signal shows that the virus mutation rate is low, as expected.
Among the 649 cases, the researchers found that the virus was introduced 277 times, and in a quarter of these, only one more case was secondarily infected before the chain was broken. A fifth of introduced cases caused an ongoing chain of spread called a transmission lineage. However, 57% of the cases did not cause even one more case.
Case Detection
The study also shows that most of the transmission lineages in NZ were from viral strains brought in from North America, which is probably a reflection of the high prevalence of the virus in that continent during the period when these cases occurred. However, the researchers did not find any sign of the virus in NZ before February 26.
The study also shows that case detection is most efficient in the second half of the epidemic than in the earlier part, because the detection lag dipped as the age of transmission lineages went down. In other words, fewer cases went undetected as the time from the first inferred transmission event in each cluster to the first detected case dropped.
Drivers of Transmission
The most significant clusters of transmission occurred after social gatherings. The biggest one is thought to have come from the US, and spread via a superspreading event (a wedding) in NZ before the lockdown, in the jurisdiction of the Southern District Health Board.
The subsequent lockdown-related change in transmission of this cluster shows how effective the intervention is. The researchers saw that the effective reproduction number Re went down from 7 at the outset to 0.2 by the end of March. Most of the cases in this cluster were sequenced in the current study, and genomic analysis threw up five hitherto unrelated cases that had escaped the first epidemiological tracing study.
Implications
The findings from this study show how important this tool can be in tracing case transmission. For one, it can help to trace unknown cases using genomic similarity. Secondly, as the researchers comment, “This cluster, seeded by a single-superspreading event that resulted in New Zealand’s largest chain of transmission, illustrates the link between micro-scale transmission to nation-wide spread.”
And thirdly, the effectiveness of the lockdown is reflected in the dramatic drop in Re in this cluster as well as the very low number of continuing infections resulting from virus importations into NZ.
At present, community transmission is absent in NZ, but border quarantine facilities continue to throw up confirmed cases because of the high case number in other places. The lesson? “Ongoing genomic surveillance is an integral part of the national response to monitor any re-emergence of the virus, particularly when border restrictions might eventually be eased.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources