An international team of researchers has conducted a study showing that differences in the human microbiome may influence the ability of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to infect host cells.
Rob Knight (University of California, San Diego) and colleagues say the study provides evidence that the human microbiome mediates the infectivity of SARS-CoV-2 by modifying a thick layer surrounding the host plasma membrane called the glycocalyx.
The researchers suggest that understanding shifts in the composition of host-microbial communities could help to improve the risk stratification of patients and potential approaches to prevention and treatment.
A pre-print version of the article is available in the server bioRxiv*, while the article undergoes peer review.
More about the glycocalyx
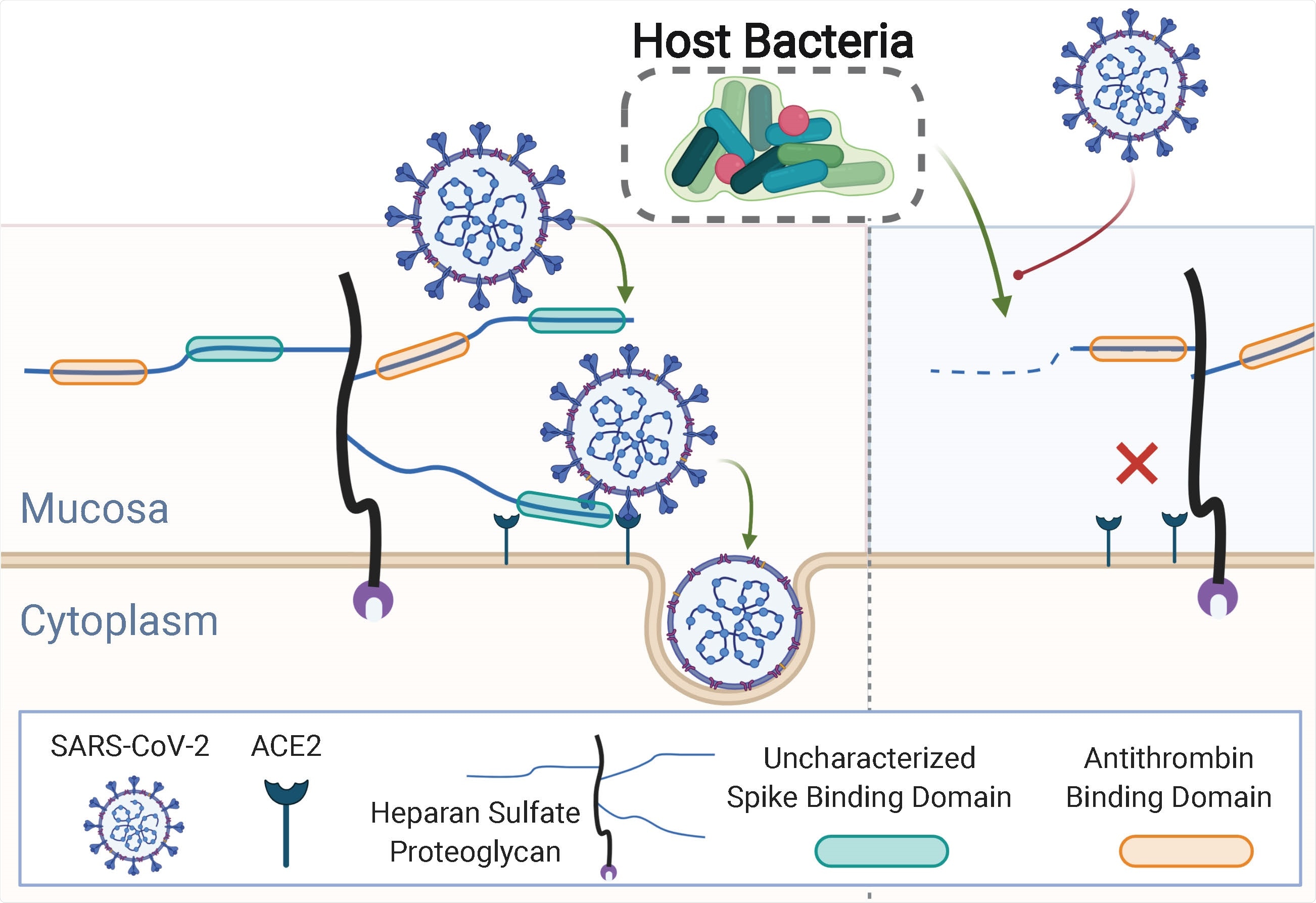
The glycocalyx is made up of a rich network of glycans and glycoconjugates that many viral pathogens bind to in order to pass through the protective glycocalyx layer and attach to protein receptors on the host cell plasma membrane.
In the case of SARS-CoV-2, a viral surface protein called “spike” targets a glycosaminoglycan (GAG) called heparan sulfate (HS). The virus requires attachment to HS in order to bind the host cell surface receptor angiotensin-converting enzyme 2 (ACE2), which enables viral membrane adhesion and host cell entry.
“Reducing the interaction between the SARS-CoV-2 spike protein and host-cell HS is, therefore, an attractive approach to reduce viral docking and subsequent infection,” write Knight and team.
What did the researchers do?
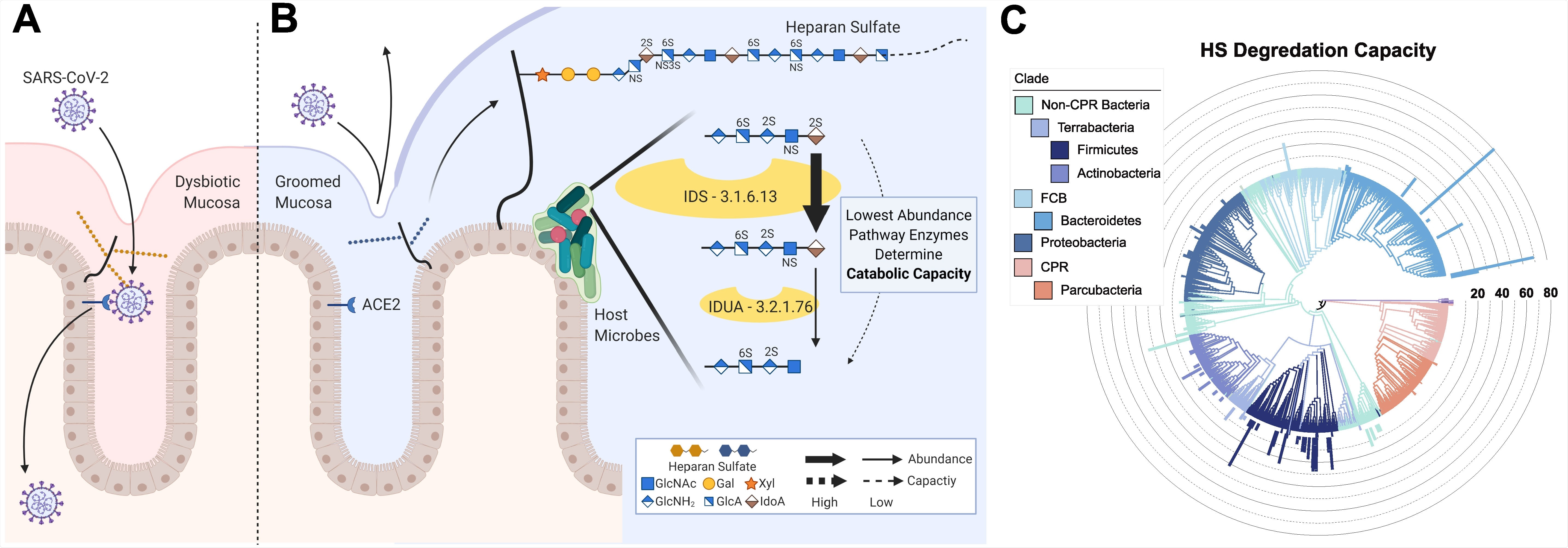
Several bacterial taxa are known to produce enzymes that modify certain types of GAG, including HS, and much research into GAGs has been advanced by using purified bacterial enzymes.
Now, Knight and colleagues have developed a computational model of HS catabolism to assess the ability of human microbes to degrade HS.
The team found that the genomes of common human-associated commensal bacteria encoded enzymes that degrade HS.
The presence of these HS-modifying bacteria was lower in bronchoalveolar lavage fluid (BALF) samples taken from patients with COVID-19 than among samples taken from healthy controls.
The researchers also found that in two independent gut microbiome datasets, levels of HS-modifying bacteria were reduced with increasing age and enhanced in females compared with males.
Older age and male gender are both known unmodifiable risk factors for developing COVID-19 following SARS-CoV-2 infection, and this finding may help to explain this increased susceptibility, says the team.
BALF RNA-seq data compared between studies. RNA-seq studies (x-axes) compared by log-ratios (y-axes) of predicted HS-modifying species relative to non-HSmodifying (A), HSase relative to housekeeping set (B), and N-acetylglucosamine-6-O-sulfatase relative to housekeeping set (C).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The HS-modifying bacteria blocked SARS-CoV-2 binding
Finally, the team showed that the HS-modifying commensals were able to prevent the binding of the SARS-CoV-2 spike protein to host cells.
“We provide evidence for a role of the human microbiome in mediating SARS-CoV-2 infectivity via modification of the host glycocalyx,” writes Knight and colleagues. “HS-modifying bacteria in human microbial communities may regulate viral adhesion, and loss of these commensals could predispose individuals to infection.”
Although the precise underlying protective mechanism is not yet known, the Bacteroide strains Bacteroides ovatus and Bacteroides thetaiotaomicron, which are highly prevalent in human gut bacteria, have previously been shown known to catabolize HS, say the researchers.
Indeed, the team went on to show that in vitro treatment of H1299 human lung cells with B. ovatus and B. thetaiotaomicron catabolized cell-surface HS and reduced SARS-CoV-2 protein binding by 20 to 30 times, compared with untreated H1299 cells.
Bacteria species encoding heparan sulfate (HS) lyase (HSase) are depleted in COVID patients compared to controls. BALF RNA-seq data from healthy subjects (control) and COVID-19 patients (COVID) (x-axes) compared by log-ratios (y-axes) of predicted HSmodifying species relative to non-HS-modifying (A), HSase relative to housekeeping set (B), and N-acetylglucosamine-6-O-sulfatase relative to housekeeping set (C). Significance was evaluated by a t-test and error bars represent the standard error of the mean. Presented p-values are from unpaired two-tailed t-test.
What are the implications of the study?
“This report provides evidence for the role of glycocalyx-microbiota interactions as a novel competitive mechanism in the fight against SARS-CoV-2,” write the researchers.
They say further work is needed to clarify the translational implications of the findings since it remains unclear which particular microbial enzymes degrade GAGs to prevent SARS-CoV-2 binding. It is also not clear whether these enzymes could be used to limit viral spread during an active infection.
Nevertheless, “the analyses presented here describe a key mechanism by which host microbiomes might play a role in SARS-CoV-2 infection, provide a potential explanation for health disparities among age groups in COVID-19 disease and highlight the importance of characterizing the potential future molecular arms race between humans and viruses,” say the researchers.
“Exploration of the vast interplay between host-microbes and viral infection could allow for improved risk stratification, prevention, and intervention,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources