Researchers in the United States have shown that failure to prevent the importation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into Massachusetts led to superspreading that drastically altered the course of the epidemic and amplified the outbreak in this major urban area.
The findings show how the virus can easily spread between seemingly disconnected groups of people and highlight the importance of prioritizing prevention, detection, and mitigation measures as part of efforts to protect public health during the coronavirus disease 2019 (COVID-19) pandemic.
The results also support the use of real-time genomic epidemiology for informing contact tracing and in helping to distinguish local outbreaks from those resulting from importation.
Bronwyn MacInnis (Broad Institute of Harvard and MIT) and colleagues say the genome sequencing approach used here could become important in helping to tackle the challenges faced as schools and workplaces start to reopen as lockdown measures are relaxed.
A pre-print version of the paper is available on the medRxiv* server, while the article undergoes peer review.
Impact of the pandemic particularly severe in Massachusetts
Since the first cases of COVID-19 were first identified in Wuhan, China, late last year, the causative agent SARS-CoV-2 has now infected more than 24 million people and caused more than 822,000 deaths.
The impact of the outbreak has been particularly severe in the state of Massachusetts, where more than 126,000 cases have now been confirmed and where almost 9,000 people have died.
The first COVID-19 case in Massachusetts was identified on February 1st, 2020. Case counts then quickly increased through the beginning of March and peaked during the third week of April.
The Boston area, which is home to about 70% of the Massachusetts population, has accounted for most (around 80%) of the infections and deaths that have occurred in the state to date.
The pandemic has disproportionally affected vulnerable populations, especially in congregate living environments such as homeless shelters, care homes, and prisons.
Superspreading played a major role
As with previous coronavirus outbreaks, a prominent feature of the COVID-19 pandemic has been superspreading events, where an infected individual transmits the virus to an unusually large number of people who then become secondary cases.
However, case counts alone cannot be used to reliably determine the extent to which an event may have contributed to overall transmission, nor can they differentiate between superspreading and other types of local and intense transmission.
“Yet understanding how the virus is actually spreading is critical for prioritizing public health interventions: cluster-based spread may be controlled with more limited restrictions than the population measures required to curb community-based transmission,” write MacInnis and team. “Genomic data can reveal connections between cases that cannot be detected through conventional epidemiology alone, including direct evidence of superspreading based on shared viral sequences.”
What did the researchers do?
The team performed genomic sequencing and phylogenetic analysis of 772 complete SARS-CoV-2 genomes acquired from nasopharyngeal samples collected between January 29th and May 9th in Boston.
MacInnis and colleagues identified more than 80 importations into the Boston area, mainly from elsewhere in the United States and from Europe, and revealed the role early superspreading events had played in driving community transmission.
The researchers studied two superspreading events that had very different impacts owing to differences in when they occurred and the populations that were involved.
What did the study find?
One event resulted in rapid spread among a vulnerable population but led to little onward spread. The other event significantly contributed to ongoing community spread, including among homeless people, and led to transmission among other domestic and international areas. This event involved a large cluster of cases related to a business conference that took place in Boston in late February.
“Ultimately, more than 90 cases were diagnosed in people associated with this conference or their contacts, raising suspicion that a superspreading event had occurred there,” said MacInnis and team.
The researchers say their analysis did indeed reveal a signature of superspreading among the conference-associated cases.
Twenty-eight genomes formed featured a particular single nucleotide polymorphism, the parent lineage of which had been widely distributed in Europe during January and February.
The researchers say one single introduction had an outsize effect on subsequent transmission due to superspreading in a highly mobile population very early on during the pandemic when many mitigation measures had not yet been put in place.
By contrast, another superspreading event that occurred in a skilled nursing facility, had little effect on onward transmission, since it occurred later and affected a more isolated population.
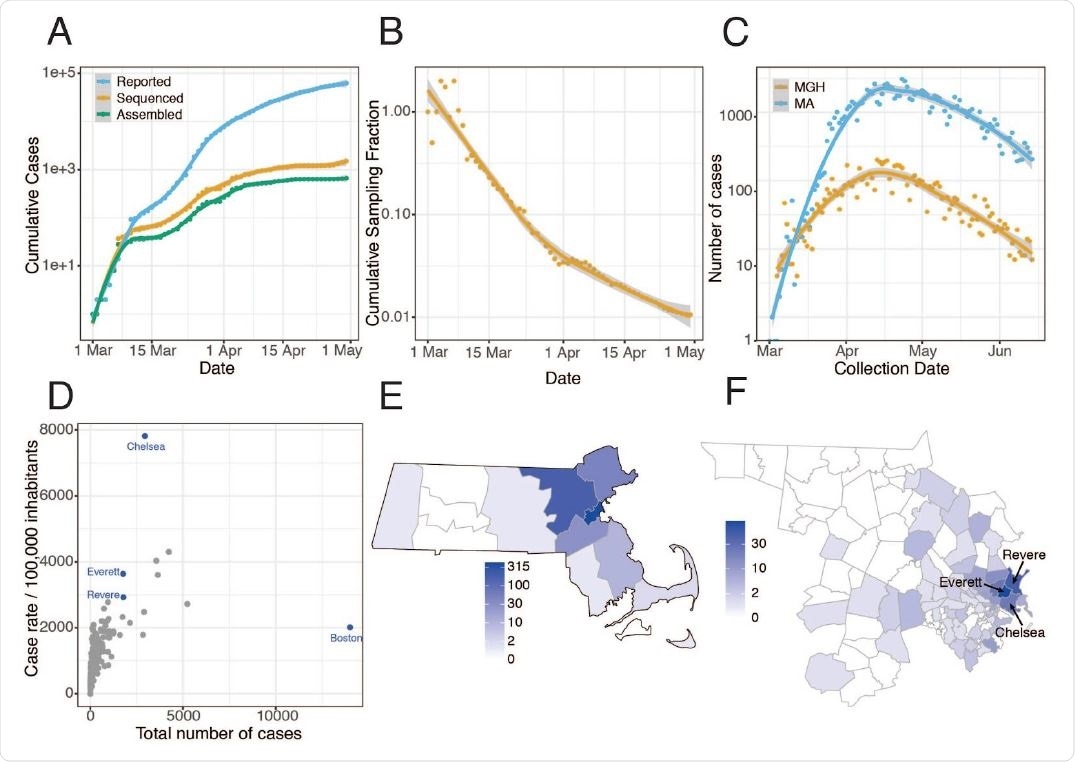
Epidemiology of SARS-CoV-2 in Massachusetts and of sequenced viral genomes. A. Cumulative confirmed and presumed cases reported state-wide in MA ( 7 ) from March 1 through May 1, 2020, and the number of these cases that were processed (orange) and successfully yielded complete genomes with >98% coverage (green) in this study. B. Cumulative proportion of all MA confirmed positive cases with complete genome sequences from unique individuals that are part of this dataset over time. C. Daily reported cases across MA from March 1 through June 15 statewide (blue) and at MGH (orange). D. Total number of cases compared to cases per 100,000 people for cities across MA. Cities in blue are highly represented in the genome dataset. E. Distribution of MA cases with sequenced viral genomes by county. F. As in E but showing only Middlesex and Suffolk counties, the two counties with the highest number of sequenced samples, by zip code. Cases associated with congregate living environments were excluded from the maps in E and F.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Real-time genomic epidemiology may be increasingly valuable
The researchers say their analysis enabled them to reassure hospital staff that it was not poor infection control that had led to the superspreading, since it showed that despite numerous introductions in the facility, only one was responsible for 90% of cases.
“Our results highlight the failure of measures to prevent the importation into MA early in the outbreak, underscore the role of superspreading in amplifying an outbreak in a major urban area, and lay a foundation for contact tracing informed by genetic data,” writes the team.
The authors suggest that “real-time genomic epidemiology may be increasingly valuable as schools and workplaces navigate the challenges of reopening, as it can help distinguish between local outbreaks within institutions and introductions from outside.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
MacInnis B, et al. Phylogenetic analysis of SARS-CoV-2 in the Boston area highlights the role of recurrent importation and superspreading events. medRxiv, 2020. doi: https://www.medrxiv.org/content/10.1101/2020.08.23.20178236v1
- Peer reviewed and published scientific report.
Lemieux, Jacob E., Katherine J. Siddle, Bennett M. Shaw, Christine Loreth, Stephen F. Schaffner, Adrianne Gladden-Young, Gordon Adams, et al. 2020. “Phylogenetic Analysis of SARS-CoV-2 in Boston Highlights the Impact of Superspreading Events.” Science, December, eabe3261. https://doi.org/10.1126/science.abe3261. https://www.science.org/doi/10.1126/science.abe3261.