Researchers at the University of California and the Lawrence Berkeley National Laboratory have made important discoveries about a protein found on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that could lay the foundations for the development of new treatments.
James Hurley and colleagues say X-ray crystallography of the viral protein - ORF8 - revealed two dimerization interfaces unique to SARS-CoV-2 – the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic.
The study identified the presence of a covalent dimer that is a recently evolved addition among beta coronaviruses and unique to SARS-CoV-2. It also revealed a non-covalent dimer that is formed by another sequence specific to SARS-CoV-2 and absent in SARS-CoV-1 – the agent that caused the 2002 to 2003 SARS outbreak.
The presence of these two interfaces may explain how the protein can form large-scale assemblies that enable the virus to suppress and evade host immune responses, says the team.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
Understanding the molecular basis of COVID-19 severity
The severity of the current coronavirus disease 2019 (COVID-19) pandemic, compared with the outbreaks caused by SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV), has triggered great efforts to elucidate the underlying molecular basis of this severity.
The viral protein ORF8 is one of the most rapidly evolving accessory proteins among the beta coronaviruses and has previously been proposed to interfere with host immune responses.
The SARS-CoV-2 ORF8 protein exhibits less than 20% sequence similarity to the ORF8 found on SARS-CoV-1 and is highly divergent.
The ORF8 proteins of both SARS-CoV-1 and SARS-CoV-2 contain a signal sequence for import into the endoplasmic reticulum (ER), where SARS-CoV-2 ORF8 interacts with a range of host proteins in the ER lumen. Many of these host proteins are involved in ER-associated protein degradation.
Hurley and colleagues say that since the presence of ORF8 is one of the main indicators of SARS-CoV-2 infections, ORF8 is presumably secreted by the ER rather than retained there.
Interfering with the host immune response
SARS-CoV-2 ORF8 has been proposed to interfere with host immune responses in several ways. Studies have shown that the ORF8 of SAR-CoV-2, but not SARS-CoV-1, downregulates the expression of major histocompatibility complex class I (MHC-1) proteins. Exogenous overexpression of ORF8 has also been shown to disrupt interferon 1 (IFN-1) signaling.
“These observations suggest the relationship between ORF8 structure, function, and sequence variation may be pivotal for understanding the emergence of SARS-CoV-2 as a deadly human pathogen,” said Hurley and team.
However, to date, no 3D structure template of any ORF8 coronavirus protein has been elucidated, and no homologs of known structures are available that have sufficient sequence identity for reliable alignment.
Another protein found on SARS-CoV-1 and SARS-CoV-2 called ORF7a is the most closely related 3D structure, but its core is around half the size of ORF8, and its primary sequence identity is negligible, say the researchers.
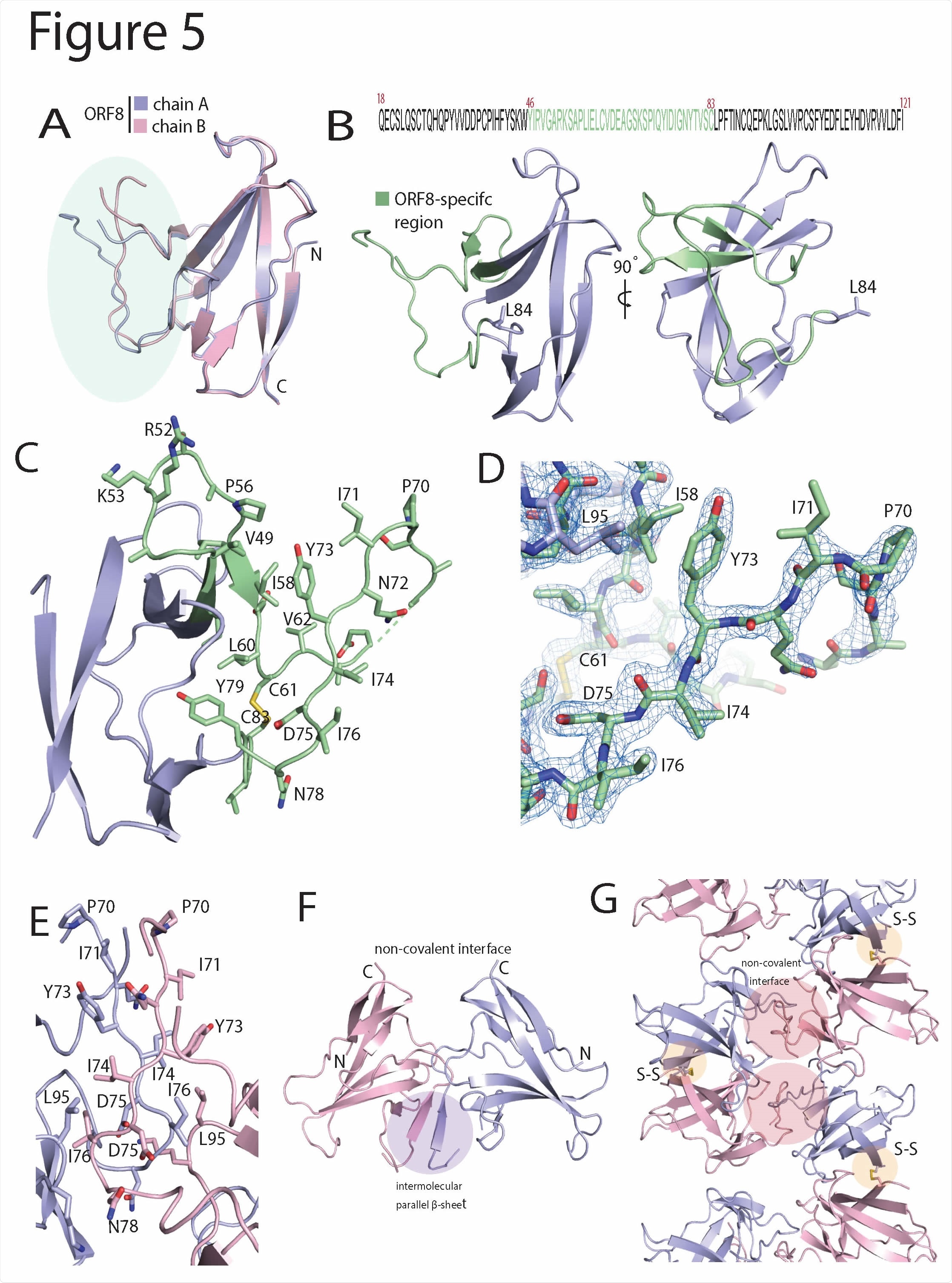
SARS-CoV-2 ORF8 contains a large, unstructured insertion. (A) Structural alignment of ORF8 chain A and B of the disulfide-linked dimer. The region corresponding to the ORF8- specific region is highlighted in green. (B) Primary sequence of the SARS-CoV-2 ORF8 construct used in this study is shown top panel. The ORF8-specific region is highlighted in green. Below is a cartoon representation of the monomer with the ORF8-specific region colored green. (C) A closeup of the ORF8-specific region is annotated. Notable residues are shown as sticks and labeled accordingly. (D) Stick representation of the insertion with 2Fo-Fc electron density map. The map is contoured at 2σ and represented as a blue mesh. (E) The crystallographic contact between ORF8 chain A and B form a non-covalent interface highlighted by an extensive array of hydrophobic residues. The residues are annotated and shown in stick form. (F) The non-covalent interface between ORF8 chain A and B forms a short, parallel β-sheet (G) Cartoon representation of alternating covalent disulfide and non-covalent interfaces in the ORF8 crystal lattice.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
What did the researchers do?
Now, Hurley and colleagues have determined the structure of SARS-CoV-2 ORF8 using X-ray crystallography.
The structure confirmed the overall similarity between the core fold of SARS-CoV-2 ORF8 and SARS-CoV-1 ORF7a.
The SARs-CoV-2 ORF8 had an approximately 60-residue core that was similar to SARS-CoV ORF7a, as well as an additional two dimerization interfaces.
“The structure reveals two novel dimer interfaces for SARS-CoV-2 ORF8 unique relative to all but its most recent ancestors in bats,” writes the team.
The study revealed a covalent disulfide-linked dimer that is formed through a SARS-CoV-2 specific sequence at the N-terminal of the protein.
“The covalent dimer is an evolutionarily recent addition among human beta coronaviruses that is unique to SARS-CoV-2,” say Hurley and team.
The analysis also revealed a distinct non-covalent dimer formed through another sequence specific to SARS-CoV-2.
The two interfaces may mediate immune evasion and suppression
“The structure shows two sequence regions unique to SARS-CoV-2 control the oligomerization and crystal packing of ORF8, and potentially mediate higher-order macromolecular assemblies unique to SARS-CoV-2,” say the authors.
The presence of these two interfaces shows how recent evolutionary changes in the sequence of SARS-CoV-2, compared with its more benign precursors, may play a role in the formation of unique large-scale assemblies that could potentially mediate unique functions in the suppression and evasion of the immune system, they explain.
“Together, our results set the foundation for elucidating essential aspects of ORF8 biology to be leveraged for the development of novel therapeutics,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hurley J, et al. Structure of SARS-CoV-2 ORF8, a rapidly evolving coronavirus protein implicated in immune evasion. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.08.27.270637
- Peer reviewed and published scientific report.
Flower, Thomas G., Cosmo Z. Buffalo, Richard M. Hooy, Marc Allaire, Xuefeng Ren, and James H. Hurley. 2021. “Structure of SARS-CoV-2 ORF8, a Rapidly Evolving Immune Evasion Protein.” Proceedings of the National Academy of Sciences 118 (2). https://doi.org/10.1073/pnas.2021785118. https://www.pnas.org/doi/full/10.1073/pnas.2021785118.