A team of Canadian scientists has recently discovered a wide-range interaction between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and host proteins using proximity-dependent biotinylation approaches. The study is currently available on the bioRxiv* preprint server.
SARS-CoV-2, the causative agent for the coronavirus disease 2019 (COVID-19) pandemic, is a deadly virus that has already claimed over 890,000 lives worldwide. To date, no specific medicine or vaccine has been identified to tackle the virus spread. To effectively design and develop therapeutic interventions, it is of utmost importance to determine the virus-host protein-protein interaction patterns, which is a prerequisite for virus survival and replication inside host cells.
.jpg)
Transmission electron micrograph of SARS-CoV-2 virus particles, isolated from a patient. Image captured and color-enhanced at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
In the current study, the scientists performed proximity-dependent biotinylation (BioID) with the miniTurbo enzyme to 27 N- and C-terminally tagged SARS-CoV-2 proteins in a lung adenocarcinoma cell line to characterize intracellular localization of viral proteins and to map virus-host protein-protein interaction network.
Why are BioID approaches better than conventional affinity purification approaches?
The SARS-CoV-2 genome contains one large 5’ open reading frame (ORF) that encodes two large polyproteins, which undergo viral protease-mediated proteolysis to generate non-structural proteins (NSP1 – NSP16). In addition, the SARS-CoV2 genome encodes structural proteins, including spike protein, viral envelope, membrane proteins, and nucleocapsid. All these proteins together form the SARS-CoV2 proteome, and the interactions between viral proteins and host proteins are the essential requirement for the virus to continue its lifecycle inside host cells.
The conventional affinity purification approaches are the golden standard for analyzing biochemically stable protein-protein interactions; however, these approaches are not very suitable for studying weak or transient interactions and interactions that occur at poorly soluble intracellular locations, such as membranes.
The proximity-dependent biotinylation approach involves the fusion of a ‘bait’ protein with a bacterial enzyme (biotin protein ligase) to facilitate covalent biotinylation of lysine residues of any proteins that are in the vicinity of the bait. Because of the covalent biotinylation, it is possible to use strong cell lysing conditions so that all cellular components (organelles, membranes, protein complexes) can be solubilize and analyze to identify interacting proteins. Afterward, all biotinylated proteins are purified using streptavidin affinity purification together with mass spectrometry.
The BioID approach used in the current study
Initially, an E. Coli BirA enzyme with a single mutation was used in BioID; however, the catalytic activity of the enzyme was relatively low. To overcome this issue, new BirA variants, such as miniTurbo, have been identified.
In the current study, the scientists conducted BioID with miniTurbo, which is an N-terminally truncated BirA protein containing many additional mutations that significantly improve the catalytic activity of the enzyme. The scientists expressed miniTurbo-tagged viral bait proteins in a human lung alveolar epithelial cell line using the lentiviral delivery system.
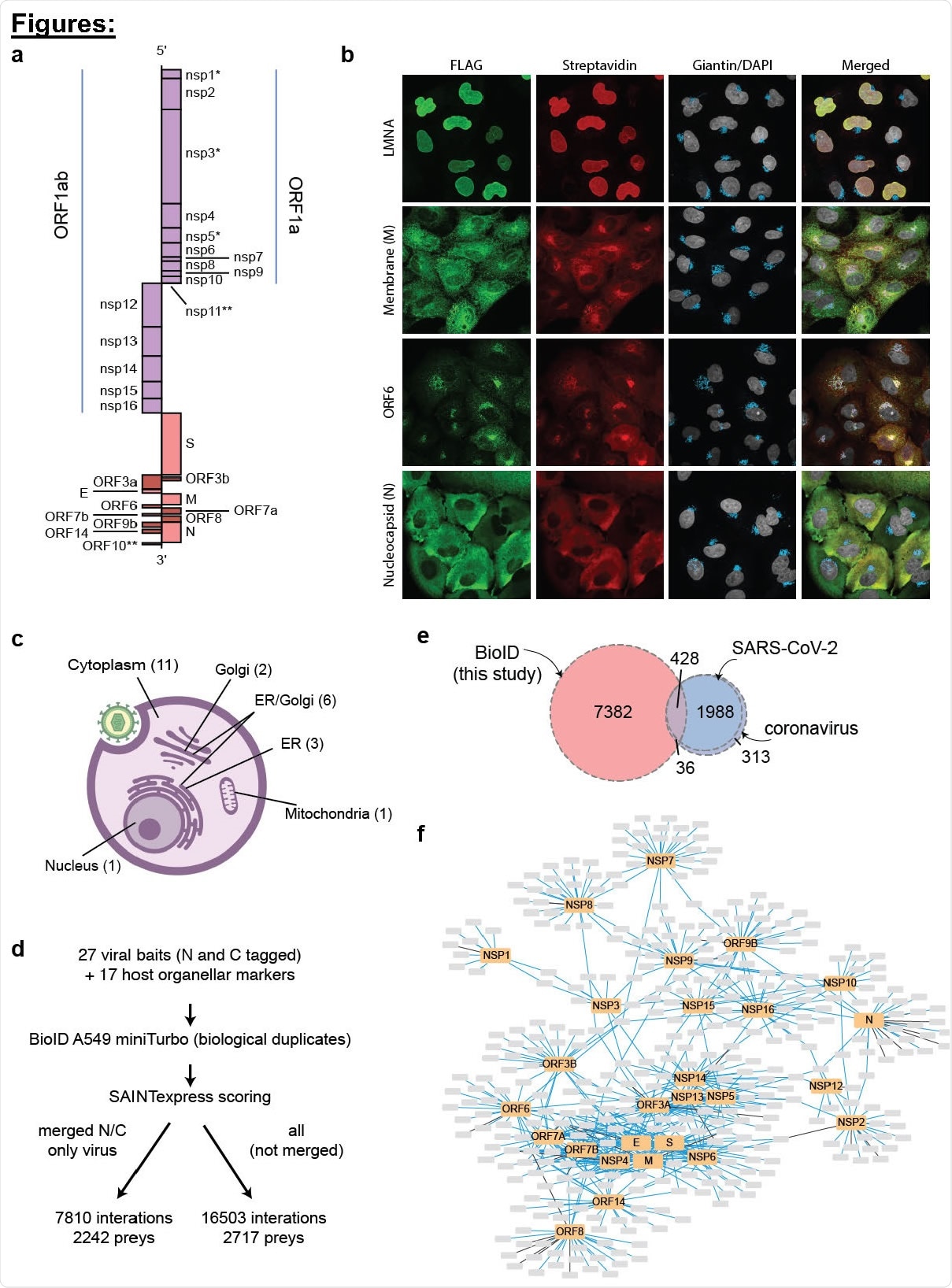
Generation of a SARS-CoV-2 - host proximity interactome in A549 cells. a, Schematic of the virus proteins profiled in this study, mapped on the genomic viral RNA (*profiled in wild type and mutant forms; **not profiled). b, Representative immunofluorescence images demonstrating miniTurbo-tagged SARS-CoV-2 proteins targeting to different subcellular compartments and miniTurbo-tagged LMNA as a representative host protein. c, Summary of immunofluorescence data (number of baits in each category indicated in parentheses. d, Overview of the dataset. e, Overlap of the proximity interactions reported here with previously reported interactions for SARS-CoV-2 or any coronavirus. f, Cytoscape representation of the most abundant (length normalized spectral counts; up to 25 are shown) proximity interactors for each viral bait see covid19interactome.org/#network for an interactive version of this image.
SARS-CoV-2 – host protein interactome
Using the miniTurbo-based BioID approach, the scientists identified proximity interactions of 7810 viral proteins with 2242 host proteins. The scientists also found that the majority of viral proteins are localized in the cytoplasm; however, many proteins are also associated with the endoplasmic reticulum, Golgi apparatus, mitochondria, and other organelle membranes.
Three viral proteins (NSP7, NSP8, and NSP12) were found to form a complex that interacts with the host proteome. The host proteins that interacted with NSP7/8 included components of the minichromosome maintenance protein complex, DNA damage checkpoint proteins, histone binding proteins, ubiquitin system proteins, proteasome components, etc. similarly, the interactome analysis of two viral proteins, NSP4 and NSP6, revealed that these proteins might be associated with membrane reorganization and viral replication organelle formation.
The viral ORF9B was found to interact with several host mitochondrial outer membrane proteins, and also with the proteins responsible for host innate immune system regulation. Regarding the utilization of host protein synthesis machinery for virion production, several viral proteins were found to interact with host ribosomal components and cytosolic RNA-binding proteins. Analysis of the viral nucleocapsid protein interactome revealed that this protein might be responsible for inhibiting host cell stress granule formation, probably by preventing the interaction between various host proteins that are essential for stress granule formation.
Study significance
The scientists created a website, namely ‘covid19interactome.org’, and uploaded the current study findings so that the entire dataset can be widely available to the scientific community. The dataset established in the current study classifies a large number of new viral-host protein-protein interactions that can be used to study the SARS-CoV-2 lifecycle in more detail, as well as to identify new therapeutic targets.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Source:
Journal reference: