A recent study by researchers at Wuhan University, China, reveals that many bat species are not potential carriers of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) because of genetic mutations in angiotensin-converting enzyme 2 (ACE2) that prevent the entry of viruses inside bat cells. The study is currently available on the bioRxiv* preprint server.
SARS-CoV-2, a deadly virus responsible for the coronavirus disease 2019 (COVID-19) pandemic, primarily transmit from human to human through respiratory droplets. However, other members of the coronavirus family, such as SARS-CoV, are known to have an animal origin. Given the similarity in genetic sequences between bat and human viruses, it has been postulated that horseshoe bats can be potential natural reservoirs of SARS-CoV-2.

Lesser horseshoe bat (Rhinolophus hipposideros). Image Credit: aaltair / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The interaction between SARS-CoV-2 spike protein and host cell receptor, ACE2, is the primary determinant of viral entry to the host cells and induction of infection. In the current study, scientists thoroughly investigated this interaction to find out the possibility of bats being the natural hosts of SARS-CoV-2.
Study design
The scientists investigated ACE2 orthologs obtained from 46 different bat species residing in rural or urban areas. Bat species from urban areas are expected to be in close proximity to humans, whereas rural bat species are expected to have minimal human contact. Overall, the experimental bat species covered about 96% of all bat species.
Important observations
After sequence alignment of bat ACE2 orthologs, they analyzed critical amino acid residues that are responsible for interacting with viral spike protein. Genetic variations were observed in all 25 critical residues that they analyzed in the study. Moreover, about 22 amino acid sites were found to be under positive natural selection; of these sites, 4 were located in the ACE2-spike protein binding region.
To study the interaction between bat ACE2 and the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein, they created stable cell lines exogenously expressing each ACE2 ortholog and recombinant fusion proteins comprising of viral RBD and human IgG Fc region. Using immunofluorescence or flow cytometry-based RBD-human Fc fusion protein-binding assay, they observed that bat ACE2 orthologs have different levels of affinity and selectivity for SARS-CoV-2. Considering this observation, the scientists suggest that not all bat species are susceptible to SARS-CoV-2 infection via ACE2-RBD binding.
To further validate their findings, the scientists developed SARS-CoV-2 spike pseudotyped viruses. They observed that the ability of bat ACE2 orthologs to support the viral entry varies between species, with some bat species showing susceptibility and some showing complete resistance to the viral entry. Experiments conducted on horseshoe bats showed that none of them support the entry of SARS-CoV-2. These observations potentially indicate that the function of ACE2 is species-dependent, and many bat species lack the ability to become natural hosts of SARS-CoV-2.
Interestingly, the scientists observed that some bat species that efficiently bind SARS-CoV-2 RBD do not get the infection, indicating that high-affinity receptor binding is not always sufficient for viral entry. Similarly, bat species that show less efficient binding to SARS-CoV-2 RBD get the infection. This indicates that some bat ACE2 orthologs may support the viral entry despite having minimal binding efficiency.
To evaluate the genetic mutations in the RBD-binding sites of bat ACE2 orthologs, the scientists selected two pairs of bat species that are phylogenetically close but have contrasting receptor binding and viral entry efficiencies. Of these four bat species, two were resistant to viral RBD binding and infection; one showed efficient RBD binding and infection, and the last one showed effective viral infection but less efficient RBD binding.
By introducing mutations in specific amino acid residues of bat ACE2 orthologs that are vital for RBD binding, the scientists observed that the substitution mutations are sufficient to reverse the function of ACE2 orthologs completely. Through structural analysis of bat ACE2 orthologs, they observed that these critical residues are located in the interface between the viral spike protein and ACE2 receptor, and any genetic alteration in these residues can significantly alter the receptor activity.
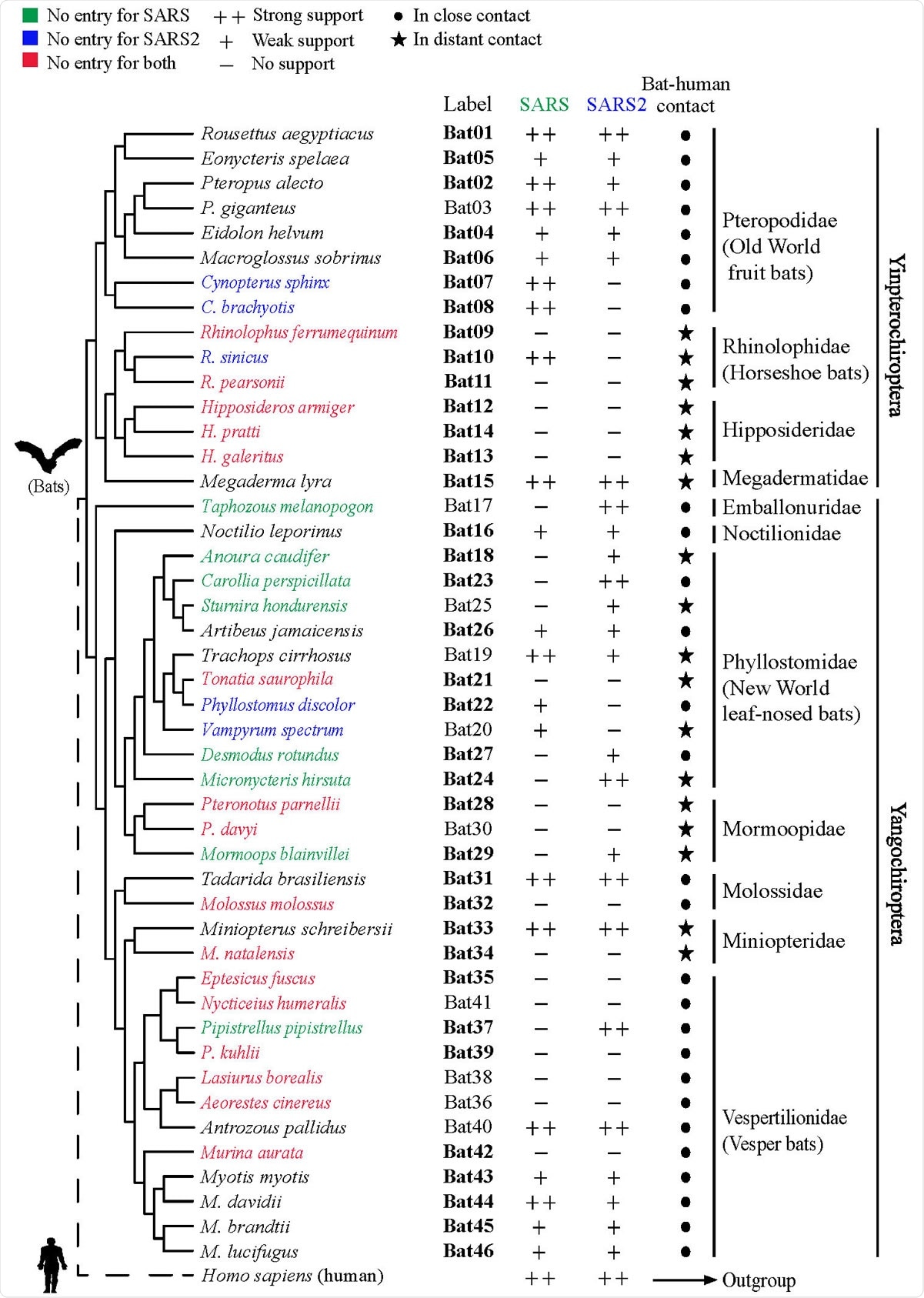
Phylogenetic tree of 46 bat species in this study. Labels of bat species in our experiments are indicated. Infection abilities of bat ACE2 to support SARS-CoV and SARS CoV-2 entry are shown with different signs: infection data are indicated as % mean values of bat ACE2 supporting infection compared with the infection supported by human ACE2; infection efficiency smaller than 5% is indicated with a minus sign (-), between 5% and 50% a plus sign (+), and greater than 50% a double plus sign (++). Labels shown in bold indicate the bat species that have been examined by in silico analyses in a recent study. Bat phylogeny was taken from previous studies.
Study significance
The current study findings provide clear evidence that the susceptibility of getting SARS-CoV-2 infection varies between different bat species. Furthermore, it is not scientifically correct to assume that all bat species can serve as potential SARS-CoV-2 reservoirs. The scientists believe that because some bat species are susceptible to human coronaviruses, humans infected with SARS-CoV-2 should restrict their activities on natural bat habitats to prevent possible zoonotic spillover.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources