More specifically, Mpro enzyme is a structurally conserved homodimer among coronaviruses, which makes it a rather attractive target for the development of antiviral therapeutics. Recently, a plethora of high resolution apo and inhibitor-bound structures of Mpro have been determined, enabling, in turn a structure-based drug design.
From the chemical perspective, Mpro is a cysteine protease characterized by a non-canonical histidine-cysteine (Cys-His) catalytic dyad. In addition to His41-Cys145, Mpro harbors a variety of histidines – including His163, His164, and His172.
The protonation states of the aforementioned histidines and the catalytic nucleophile Cys145 have been discussed in previous research studies on SARS-CoV-1 Mpro (which causes original SARS), but not for SARS-CoV-2. However, given the pressing need for efficacious treatment options in our fight against COVID-19, the quest for Mpro inhibitors has been intensified.
For example, several notable inhibitors have been identified, and their crystal structures released, such as alpha-ketoamides and peptidomimetic N3. These inhibitor-bound structures are indeed unique starting points for further drug optimization strategies.
In this new study, a research group led by James C. Gumbart of the Georgia Institute of Technology in Atlanta, aimed to determine the structural stability of SARS-CoV-2 Mpro as a function of the protonation assignments for histidine and cysteine residues. The research was a collaborative effort between Georgia Institute of Technology, the University of L’Aquila, CNR Institute of Nanoscience, the University of Tennessee, Knoxville, the Université de Lorraine, the University of Illinois at Urbana-Champaign, Oak Ridge National Laboratory and Novartis Institutes for BioMedical Research.
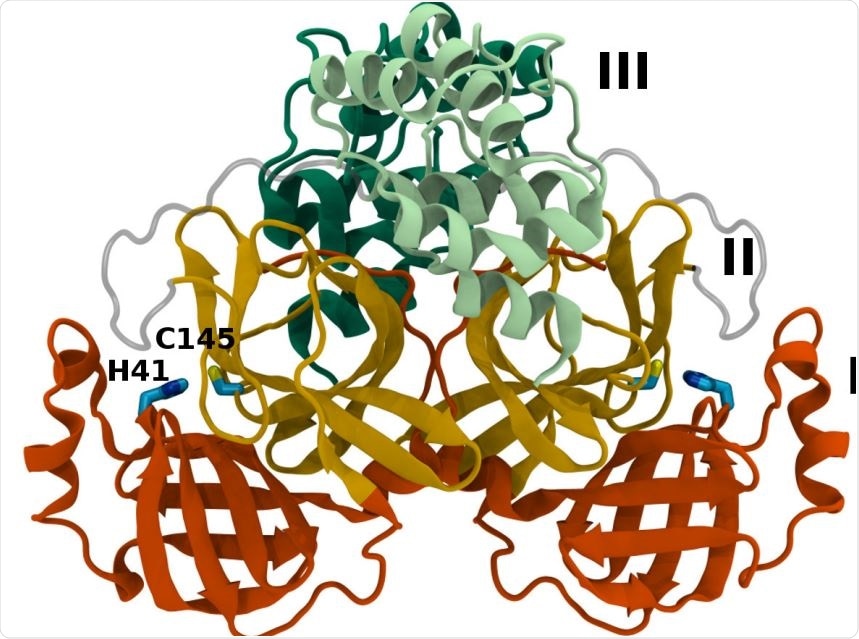
Mpro dimer structure and binding site interactions (PDB entry 6WQF). Mpro homodimer with the three domains illustrated and color coded as follows: Domain I (dark orange), domain II (gold), and domain III (light green/dark green monomer A/B) with the catalytic dyad residues, His41 and Cys145 (rendered in licorice).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Methodological approach
"We investigated the effects on the structural properties of the apo and ligand-bound systems by altering the protonation state of the catalytic dyad, Cys145 and His41, as well as those of three histidines near the substrate binding site, His163, His164, and His172", study authors summarize their methodological approach.
Taking into account kindred ambiguity in the protonation states of SARS-CoV-2 Mpro, the researchers have looked into the stability of 12 possible protonation states of the protease. Detailed molecular dynamics simulations of apo and inhibitor-bound Mpro dimers were performed for each state, with subsequent analysis of the resulting structural and dynamical properties.
Hydrogen bonding was assessed for relevant residues of the protein, as well as between the bound inhibitors and the protein. Finally, two inhibitors were studied in detail: peptidomimetic N3 and a very potent alpha-ketoamide.
Multiple protonation combinations
This study found that the protonated His41/deprotonated Cys145 state of the catalytic dyad is actually unstable in the crystal structure conformations, as evidenced by increased root-mean-square deviation, changed hydrogen bonding patterns, and disentangling of the inhibitors. Nonetheless, this state may exist as a transient reaction intermediate.
The researchers have also shown that His163 and His172 protonation states result in substantial perturbations to several hydrogen bonds in comparison to crystal structure conformations.
Additionally, decreased pocket volume was regularly observed in the simulations for the HP163 state. At the same time, free-energy calculations showed decreased affinity for both tested inhibitors in this state, which qualitatively traces the observed hydrogen bonding reductions.
In contrast to the other histidines, this research group has determined that multiple protonation combinations are viable for the His41-His164 pair. Therefore, these residues may take on different protonation states during the protein cleavage process.
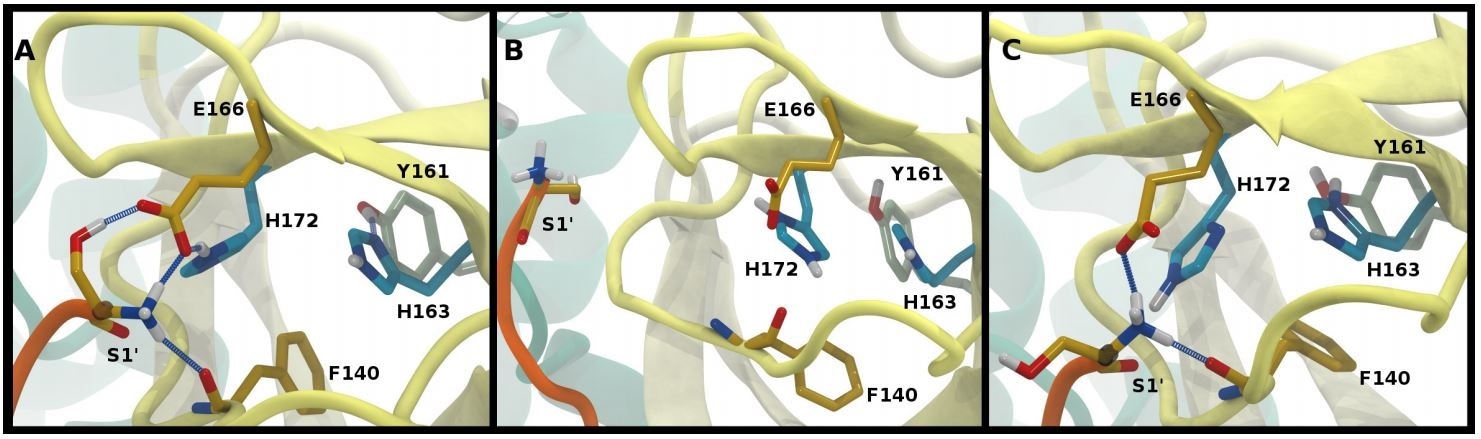
Hydrogen bonding in the S1 pocket. Example configurations of A) HD41-HE164 characteristic of robust S1 pocket interactions, B) HE41-HD164-HP172 illustrating the rupture of the S1’-Glu166 interaction and loss of the His163-Tyr161 hydrogen bond, and C) HE41-HP163-HD164 depicting the loss of the Tyr161 hydrogen bond donation and the His172-Glu166 interaction.
Implications for drug improvement and design
"Our results illustrate the importance of using appropriate histidine protonation states to accurately model the structure and dynamics of SARS-CoV-2 Mpro in both the apo and inhibitor-bound states, a necessary prerequisite for drug-design efforts", summarize study authors in this bioRxiv paper.
In other words, the most important protonation states of Mpro histidines have to be considered for optimization efforts of N3 peptidomimetic and alpha-ketoamide, as well as for the rational and targeted design of other inhibitors.
Nonetheless, in silico high-throughput screens of new potential inhibitors will benefit from being performed on each of the protonation states considered in this paper, primarily to make it feasible when ligand is present. This will definitely be a recurring theme in further research endeavors on this topic.
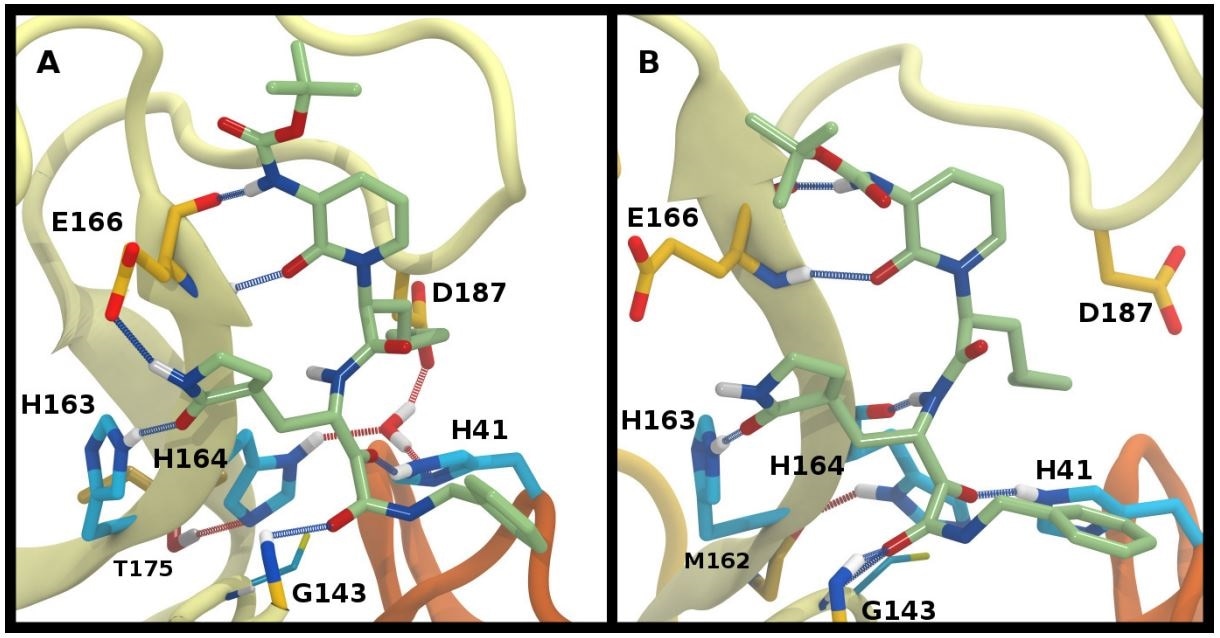
Ketoamide hydrogen bonding in the HE41-HD164 protonation state. In both panels, hydrogen bonds between the ligand (light green licorice) and the protein are indicated with a blue line, while those with water or between protein residues are red. A) Region around the crystallographic water. B) conformation in which the His164 has rotated, making a hydrogen bond with the backbone of Met162. The crystallographic water has been released.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources