The coronavirus disease 2019 (COVID-19) pandemic has become an unprecedented challenge to global health. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus causing this disease. It is a single-stranded RNA virus that binds to host cells with its trimeric spike glycoprotein S, which has two subunits. The S1 subunit facilitates receptor binding, and S2 is responsible for membrane fusion.
During infection, the receptor-binding domain (RBD) of the S1 subunit directly binds with the human angiotensin-converting enzyme 2 (ACE2) receptor. RBD is shown to be the common binding site of neutralizing antibodies from convalescent patients. Thus, the RBD can serve as a key target for drugs that inhibit viral infection.
Engineering ACE2 to enhance its affinity to RBD
RNA viruses have high mutation rates and evolvability that help them acquire anti-viral drug resistance. More and more evidence shows that neutralizing antibodies are effective in fighting COVID-19. Monoclonal antibodies taken from convalescent COVID-19 patients have been shown to have high potency in neutralizing viruses. However, spike gene mutation can cause SARS-CoV-2 adaptation to such antibodies.
Similar to the anti-RBD antibodies, the extracellular domain of ACE2 can also serve as a decoy receptor to neutralize SARS-CoV-2. The therapeutic potency of ACE2 against COVID-19 has been confirmed by a few studies. Also, fusing sACE2 to the human IgG1 Fc region has been shown to increase neutralization capacity and boost pharmacokinetics to human IgG levels in mice. Increasing the affinity of ACE2 to RBD is crucial for adequate protection against viral mutation.
In a study published in the preprint server bioRxiv*, researchers from the Kyoto Prefectural University of Medicine and Osaka University Japan, discuss how they engineered ACE2 to enhance its affinity to RBD in human cells. The team of researchers introduced random mutations in the protease domain, having the interface to the RBD. Full length ACE2 mutant library expressed in 293T cells was incubated with fluorescence-labeled RBD.
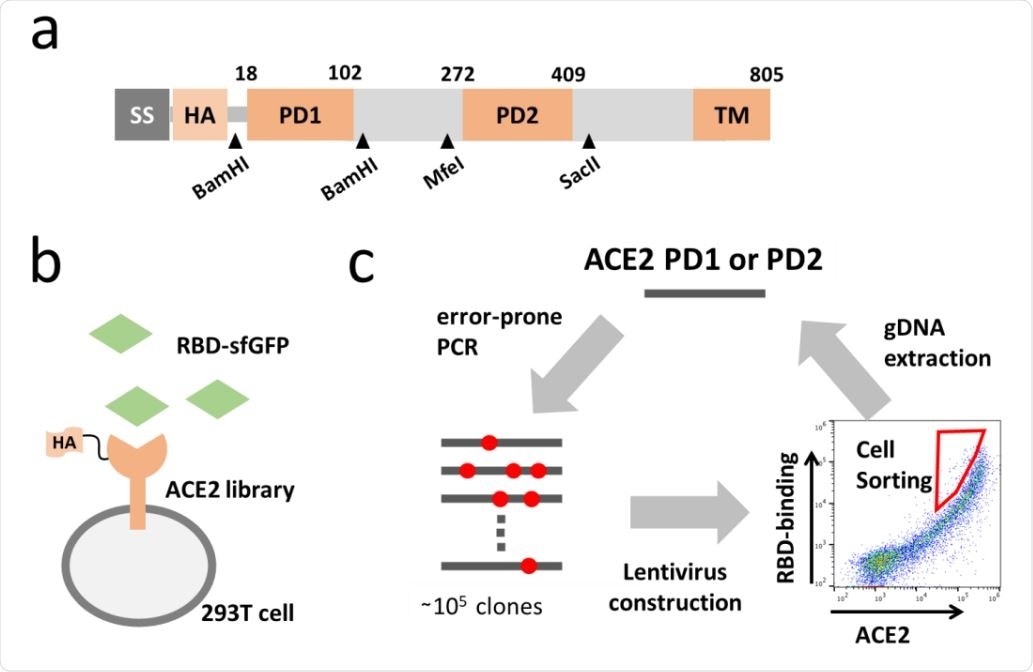
Directed evolution of ACE2. (a) Full length ACE2 was optimized to fit screening. Synthetic signal sequence and HA tag were fused to mature ACE2 and restriction sites were introduced by optimizing codon optimization for the mutated fragment replacement. (b) ACE2 mutant library was expressed in 293T cell and incubated with the RBD of SARS-CoV-2 fused to superfolder GFP (sfGFP). (c) Error-prone PCR amplification of ACE2 protease domain induced random mutations in the rate of one mutation per 100bp and generated a library of ~105 mutants. Mutant library-transduced cells were incubated with the RBD-sfGFP. Top 0.05 % population with high level of bound RBD-sfGFP relative to ACE2 expression was sorted and underwent DNA extraction, followed by next cycle mutagenesis.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Cells that showed high binding activity were sorted, and DNA extracted from these cells were further induced with random mutations for the next selection cycle. Three such cycles of random mutation and cell sorting produced ACE2 that had over 100-fold higher affinity to RBD compared to wild-type ACE2. This protein engineering system generated a virus-neutralizing drug with high affinity comparable with that of antibodies and can provide a solution to drug resistance due to escape mutation.
“Most importantly, when the neutralization potential against the authentic SARS-CoV-2 in TMPRSS2- expressing VeroE6 cells was evaluated, wild-type sACE2-Fc showed no efficiency even at 100μg/ml, whereas 3N39 sACE2-Fc demonstrated significant neutralizing effect in 6.3μg/ml.”, says the team.
The extracellular domain of the engineered ACE2 when fused to the human immunoglobulin IgG1 Fc region had a stable structure and was able to neutralize SARS-CoV-2 pseudotyped lentivirus and authentic virus at over 100-fold lower concentration than wild-type ACE2.
High affinity ACE2 decoy receptors can neutralize SARS-CoV-2
Engineering decoy receptors with improved affinity has been previously reported in cancer-related molecules. These decoy receptor drugs are used to neutralize various cytokines such as vascular tumor necrosis factor-alpha, endothelial growth factor, and CTLA-4 and are approved for the treatment of rheumatoid arthritis and orbital vascular diseases.
Though recombinant sACE2 or sACE2-Fc fusion protein has the ability to neutralize the SARS-CoV-2 virus, due to its modest binding affinity, a higher dose is required than a monoclonal antibody. The mutant ACE2s developed by the team not only had affinity comparable to anti-spike antibodies, but they also had a more extensive Interface to the RBD compared to that of antibodies, which increases their efficacy.
“We developed the screening system based on the cycle of random mutation and sorting of high-affinity population in 293T cells followed by validation of neutralizing activity in a soluble form.”
Based on the findings of their study, the team concluded that engineering decoy ACE2 receptors with directed evolution could be an effective approach in the development of a SARS-CoV-2 neutralizing drug that has an RBD affinity comparable to monoclonal antibodies, yet resist escape mutation of the virus.
According to the team, high-affinity, engineered ACE2 fused with Fc protein is a promising approach to neutralizing the SARS-CoV-2 virus. Also, the system they have developed can rapidly generate therapeutic candidates effective against many different viral diseases and may help fight future pandemics caused by viruses.
“The time frame for running one cycle of mutagenesis and sorting was just one week in our system, and we succeeded in developing optimized mutants in a couple of months without depending on patients derived cells or tissues.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
High affinity modified ACE2 receptors prevent SARS-CoV-2 infection Yusuke Higuchi, Tatsuya Suzuki, Takao Arimori, Nariko Ikemura, Yuhei Kirita, Eriko Ohgitani, Osam Mazda, Daisuke Motooka, Shota Nakamura, Yoshiharu Matsuura, Satoaki Matoba, Toru Okamoto, Junichi Takagi, Atsushi Hoshino bioRxiv 2020.09.16.299891; doi: https://doi.org/10.1101/2020.09.16.299891, https://www.biorxiv.org/content/10.1101/2020.09.16.299891v1