ACE2 is the human host receptor that enables cellular entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent that causes coronavirus disease 2019 (COVID-19).
Computational chemist and biophysicist, Rommie Amaro (University of California), and colleagues say the insights the findings provide regarding the mechanisms of viral binding and cellular entry could aid the rational design of effective therapies for SARS-CoV-2.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
SARS-CoV-2 binding to ACE2 is the initial step for viral entry
Since the first cases of SARS-CoV-2 infection were first identified in Wuhan, China, last year, the virus has swept the globe, infecting tens of millions of people and posing a significant threat to human health.
Although various efforts are underway to develop antiviral therapies and vaccines, much remains unknown about the mechanisms underlying the binding of SARS-CoV-2 to ACE2, the main interaction that enables viral entry into the host cell.
ACE2 is a homodimer protein comprising a large extracellular head domain, a small transmembrane domain, and a small intracellular tail.
The large head is further divided into a peptidase domain and a neck domain, where most of the homodimer interactions appear to occur. The neck domain is also connected to the transmembrane domain via a linker.
SARS-CoV-2 binds ACE2 using the spike glycoprotein
Both SARS-CoV-1, which caused the 2002 to 2003 SARS outbreak, and SARS-CoV-2 bind to ACE2 using a large surface viral structure called the spike glycoprotein.
When the spike protein’s receptor-binding domain (RBD) is in the “up” confirmation, it binds the peptidase domain of ACE2 with high affinity. The resulting ACE2-RBD complex consists of two heterodimers, with each polypeptide chain of the ACE2 homodimer bound to one spike RBD.
The binding of the RBD to the peptidase domain on ACE2 triggers a series of conformational changes in the spike protein that enable it to fuse to the host cell membrane and gain viral entry.
The spike glycoprotein and the complex it forms with ACE2 are, therefore, key targets in efforts to develop drugs that might curtail the unprecedented transmissibility of SARS-CoV-2.
Structural and biophysical studies of the spike RBD-ACE2 complex have pointed to structural features that likely account for the more potent affinity and infectivity of SARS-CoV-2, compared with SARS-CoV-1, and have revealed important dynamics at the RBD-PD interface.
Furthermore, some recent cryogenic electron microscopy and computational studies have indicated a significant degree of flexibility at the stalk of the spike protein and at the ACE2-RBD interface, suggesting a need to examine these complexes in the context of the cell surface, rather than under static, cryogenic experimental conditions.
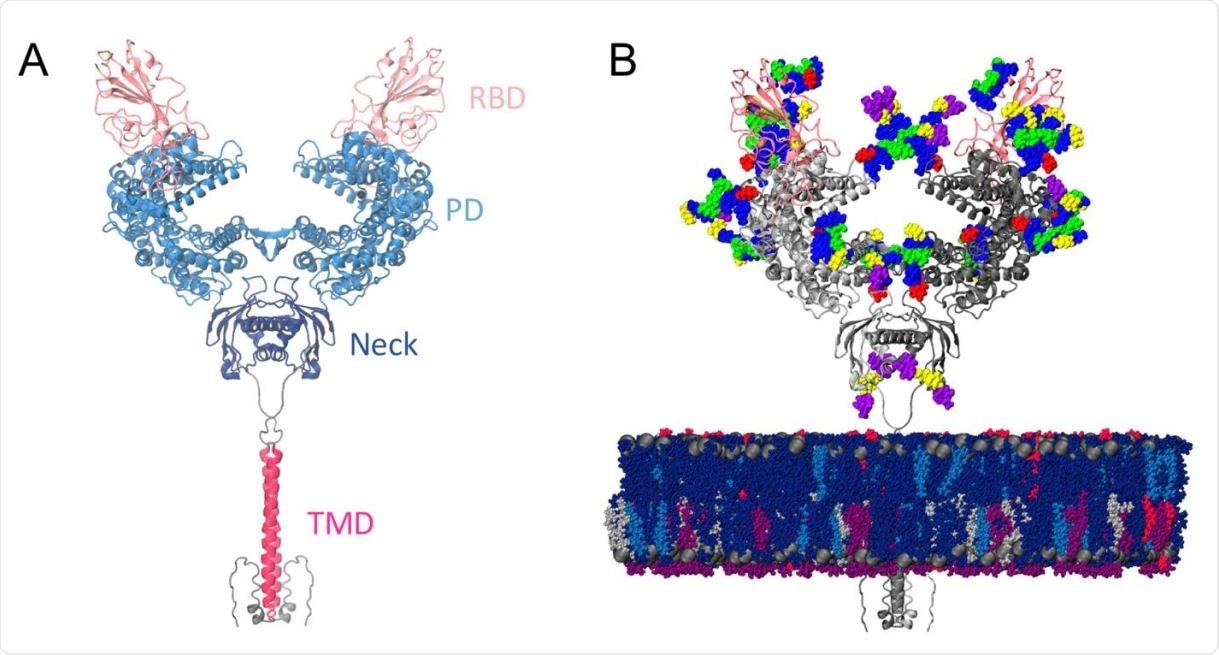
Model structure. (a) Full-length ACE2 homodimer protein structure in complex with spike protein RBDs. ACE2 peptidase, neck and transmembrane domains are shown with cartoons highlighted in blue, navy and magenta, respectively. Spike RBDs are depicted with pink cartoons. (b) Fully glycosylated and membrane-embedded model. ACE2 and RBDs are represented with gray and pink cartoons, respectively. Atoms of N-/O-glycans are shown with per-monosaccharide colored spheres, where GlcNAc is highlighted in blue, mannose in green, fucose in red, galactose in yellow, and sialic acid in purple. Lipid heads (P atoms) are represented with grey spheres, whereas lipid tails are depicted with a licorice representation using the following color scheme: POPC (navy), POPI (violet), POPE (silver), CHL (blue), PSM (magenta).
What did the current study involve?
Now, Amaro and team have performed all-atom molecular dynamics simulations of the full-length, membrane-embedded ACE2 receptor, both while unbound by spike RBD and while in complex with RBD, to probe the molecular dynamics of ACE-2 flexibility on the host receptor side.
The simulations demonstrated exceptional plasticity in the full-length ACE2 homodimer due to highly flexible hinge motions in the head-transmembrane domain linker region and profuse helix mobility in the membrane.
Remarkably, this flexibility did not disrupt the ACE2 homodimer or the ACE2-RBD interfaces. The homodimer interface remained stable at the neck domains and at the ACE2-RBD contacts, despite the dramatic motions, thereby emphasizing the high affinity of the interaction, says the team.
What are the implications of the study?
The researchers say this flexibility gave rise to a varied range of ACE2 conformations that are distinct from those seen in experimental structures and that these conformations could sterically accommodate binding of the spike protein to more than one ACE2 homodimer.
This ACE2 flexibility indicates a mechanical contribution to the spike conformational changes that are required for cellular fusion and infection, they say, and “suggests the structural basis for the possibility of finding two or more ACE2 complexes bound to the same S [spike] glycoprotein with two or more ‘RBD-up’ conformations.”
“Taken together, our findings shed further light onto the mechanisms of viral binding and cell entry required for rational design of effective SARS-CoV-2 therapeutics,” concludes the team.