The team identified important residues in the spike receptor-binding domains (RBDs) of SARS-CoV-2, the bat coronavirus RaTG13, and the pangolin coronavirus PCoV_GX that underlie the differences in activities of these spike proteins and their ability to bind to and infect host cells.

Image Credit: 2630ben / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Xinquan Wang and colleagues suggest that five residues, in particular, are critical to the evolution of the SARS-CoV-2 spike RBD, due to the role they play in enabling tight binding to the human host cell receptor angiotensin-converting enzyme 2 (hACE2).
"These results collectively indicate that strong RBD-ACE2 binding and efficient RBD conformational sampling are required for the evolution of SARS-CoV-2 to gain highly efficient infection," writes the team.
A pre-print version of the paper is available on the serve bioRxiv*, while the article undergoes peer review.
Cross-species transmission of coronaviruses an ongoing threat
Animal-to-human (zoonotic transmission) of coronaviruses represents a significant threat to human health globally, as evidenced by the emergence of SARS-CoV-1, Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 over the last two decades.
Current evidence suggests that similarly to SARS-CoV-1 and MERS-CoV, SARS-CoV-2 probably originated in bats before evolving in intermediary hosts and then jumping to humans.
Coronavirus RaTG13, which was detected in the horseshoe bat, has been identified as sharing the most significant sequence identity (96.2%) with SARS-CoV-2, thereby pointing to the likely origin of SARS-CoV-2 in bats.
Another Malayan pangolin coronavirus (PCoV) identified in China's Guangxi (GX) is also closely related to SARS-CoV-2. Genome sequencing of this virus, PCoV_GX, has also indicated a high level of shared sequence identity (85.5%) with SARS-CoV-2.
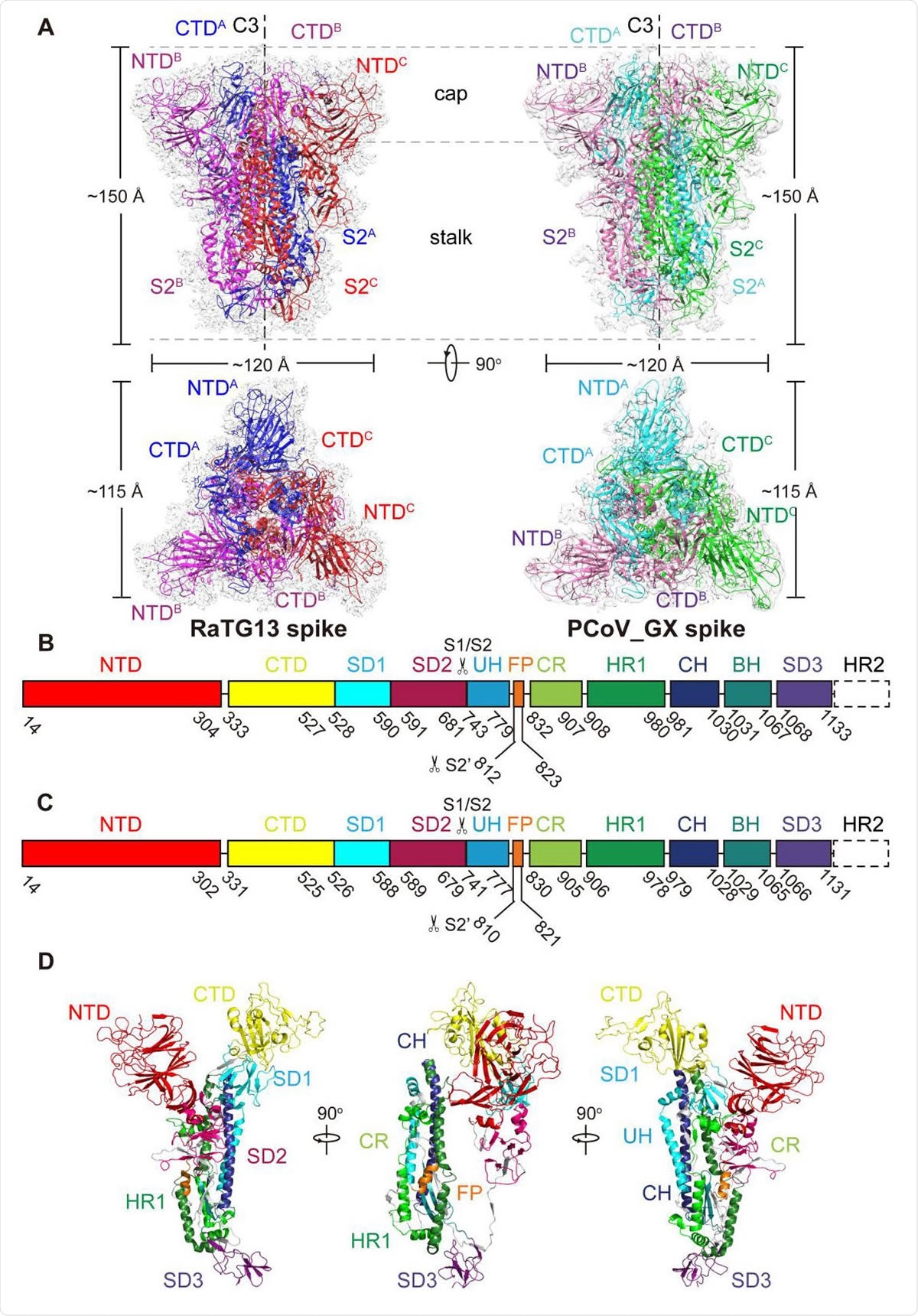
Over all structures of the RaTG13 and PCoV_G 553 X spike glycoproteins. (A) Overall structures of RaTG13 and PCoV_GX spike glycoproteins shown in side view (upper panel) and top view (lower panel). Three monomers of the RaTG13 spike are colored magenta, red, and blue, respectively; three monomers of the PCoV_GX spike are colored hot pink, green and cyan, respectively. The cryo-EM maps are shown as a semitransparent surface. The trigonal axes are shown as black dashed lines. Visible segments of each monomer are labeled accordingly. The cap and stalk parts are partitioned by gray dashed lines. (B) Schematic representation of the RaTG13 spike monomer structural domains. The domains of RaTG13 are shown as boxes with the width related to the length of the amino acid sequence. 562 The start and end amino acids of each segment are labeled. The position of the S1/S2 and S2’ cleavage sites are indicated by scissors. NTD, N-terminal domain; CTD, C-terminal domain; SD1, subdomain 1; SD2, subdomain 2; UH, upstream helix; FP, fusion peptide; CR, connecting region; HR1, heptad repeat 1; CH, central helix; BH, β-hairpin; SD3, subdomain 3. (C) Schematic representation of the PCoV_GX spike monomer structural domains. The abbreviations of elements are the same as in B. (D) Cartoon diagrams depicting three orientations of the spike monomer colored as in B and C. As the RaTG13 and PCoV_GX spike monomers have extremely similar structures, thus only the RaTG13 spike monomer was used to show the detailed architecture.
The role the spike trimer plays in cross-species transmission
As the main viral structure enabling coronaviruses to infect host cells, the role the spike trimer protein plays in cross-species transmission and infection is of major interest to researchers.
"Coronavirus spike glycoproteins recognize their host cellular receptor and mediate membrane fusion for entry, thereby functioning as the most critical coronavirus protein in determining viral evolution and cross-species transmission," say Wang and colleagues.
Cryogenic electron microscopy (cryo-EM) studies have previously shown that similar to the spike trimer of SARS-CoV-1, the SARS-CoV-2 spike trimer needs to have at least one RBD in an "up" conformation in order to bind hACE2.
"Therefore, a spike trimer with all three RBDs' down' is in a receptor-binding inactive state, and the conformational change of at least one RBD from 'down' to 'up' switches the spike trimer to a receptor-binding active state," explain the researchers.
What did the current study involve?
Now, Wang and colleagues have determined the cryo-EM structures of the spike proteins from RaTG13 and PCoV_GX spikes and compared them to the spike of SARS-CoV-2.
The analysis revealed that the RBDs of RaTG13 and PCoV_GX spikes closely resembles that of the SARS-CoV-2 spike.
All three RBDs of the RaTG13 and PCoV_GX spike trimers were in the "down" conformation, suggesting that these RBDs tend to adopt the receptor-binding inactive state.
However, on performing surface plasmon resonance experiments, the researchers found that the PCoV_GX spike RBD exhibited a similar binding affinity for hACE2 to that of the SARS-CoV-2 spike RBD. At the same time, the RaTG13 RBD demonstrated far weaker hACE2 binding.
Variations at RBD residues accounted for the variation
Next, the team identified variations at six residues in the RBD that seemed to account for these differences in hACE2 binding between the three viruses.
The residues Y449, Q493, Q498, N501, and Y505 were particularly important since they clustered together to form a patch on the SARS-CoV-2 RBD that strongly interacted with hACE2.
The researchers also pinpointed amino acid changes at two positions (Y449 and Y505) that only occurred in the RaTG13 spike RBD and not the PCoV_GX spike RBD, which the researchers say may account for the weaker binding of hACE2 by RaTG13.
"We further propose that the patch containing Y449, Q493, Q498, N501, and Y505 plays a critical role in the evolution of the SARS-CoV-2 RBD, promoting especially tight binding to hACE2 and impacting the varying affinities observed between the RBD and ACE2 orthologs in wild and domestic animals," write Wang and colleagues.
The team also identified three N-linked glycosylation sites in the spike RBD of RaTG13 and PCoV_GX, one of which (N370) is not a glycosylation site in the spike RBD of SARS-CoV-2.
"The absence of glycans linked to N370 may contribute to the more flexible RBDs of the SARS-CoV-2 spike," suggest the researchers.
They say this hypothesis is supported by another study showing that mutation of N165 in SARS-CoV-2 gave rise to an increase in the "up" conformation of RBDs, suggesting that glycans serve as a conformational control element of the RBD.
What did the authors conclude?
"Based on all these results, we propose that the tight RBD-hACE2 binding we observed is the most critical factor in determining the varied cell-entry efficiency among RaTG13, PCoV_GX, and SARS-COV-2," say the researchers.
"This and the RBD' down' to 'up' conformational change are both required for the evolution of SARS-CoV-2 to gain highly efficient transmission capability," concludes the team.
Source
Wang X et al. Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.09.21.307439

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources