In the viral genome landscape, to understand what is happening, it is vital to analyze mutations and detect recombinant forms and other anomalies among an extensive set of sequences. Scientists apply a new algorithm, Bolotie, to a large collection of SARS-CoV-2 genomes, to efficiently detect mutations and analyze recombination events. They identify multiple unique cases of recombination between 4 prominent clades of the virus.
The current pandemic caused by a newly emerged strain has brought the world to a halt. As of 22 September 2020, over 31.76 million confirmed cases due to coronavirus disease 2019 (COVID-19) worldwide are reported, and the death toll has exceeded 973,000. Caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a strain of β-coronavirus (CoV). The precise origin of this strain is not yet known; it is hypothesized to have emerged as a result of a recombination event between a bat and a pangolin. Community transmission of the virus, as well as anti-viral treatments, can initiate mutations in the virus. These mutations may result in more virulent strains that may cause higher mortality rates, or be resistant to treatments*.
While the genetic diversity of the SARS-CoV-2 has been increasing slowly compared to other RNA viruses, coronaviruses are known to undergo mutations at high rates just as other RNA viruses. Recombination events, such as inter- and intra-host, are well known to occur frequently. At present, 5 to 7 major circulating clades of SARS-CoV-2 are identified based on multiple variants common to large numbers of isolates in the GISAID database.
The low divergence of near-identical genomes that are sequenced in short periods presents a statistical challenge that is not addressed by methods available currently. In a recent bioRxiv** paper, Ales Varabyou et al. from John Hopkins University present an efficient method designed to detect recombination and reassortment events between clades of viral genomes – a new algorithm called ‘Bolotie’.
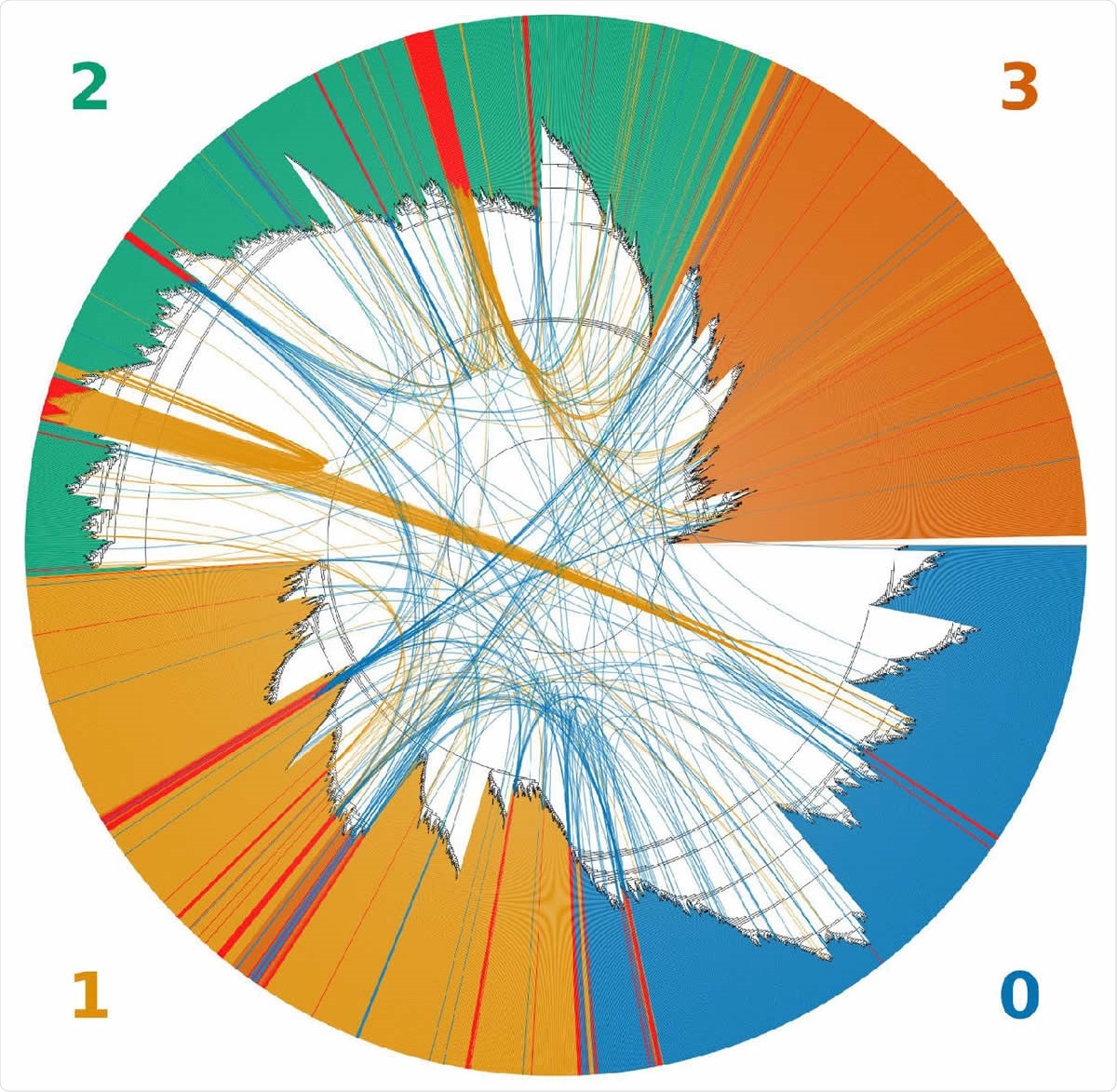
An unrooted topological cladogram of 4,249 SARS-CoV-2 genomes including 225 recombinants labeled as red bars. Arcs link each recombinant to both inferred parental genomes. The color of the arc corresponds to the color of the clade to which a recombinant was clustered within the tree. Clades correspond to the GISAID clades GR (0), GH (1), G (2) and all minor lineages combined (4).
It is critical to scrutinize the sequenced genomes of SARS-CoV-2 for both novel mutations and recombinations to develop effective treatments and successful vaccines. The method employed for this survey needs to be: rapid in performing the analysis (for extensive data - nearly 100,000 genomes to date that has been generated for SARS-CoV-2); less intensive computationally; demanding fewer resources; highly efficient. Currently, for SARS-CoV-2 genomes, there are 512 trillion unique triplets of sequences available; performing a similarity analysis for each triplet is computationally infeasible.
The authors have presented methods that are designed such that the novel sequences are analyzed efficiently without the need to rerun the entire protocol. The analysis, including alignment and index construction, of the 87,695 genomes using Bolotie took a total of ~5.5 hours using 36 threads on a two 10 core Intel Xeon E5-2680 v2 processors; analyzing a single additional genome took on average only ~30 seconds.
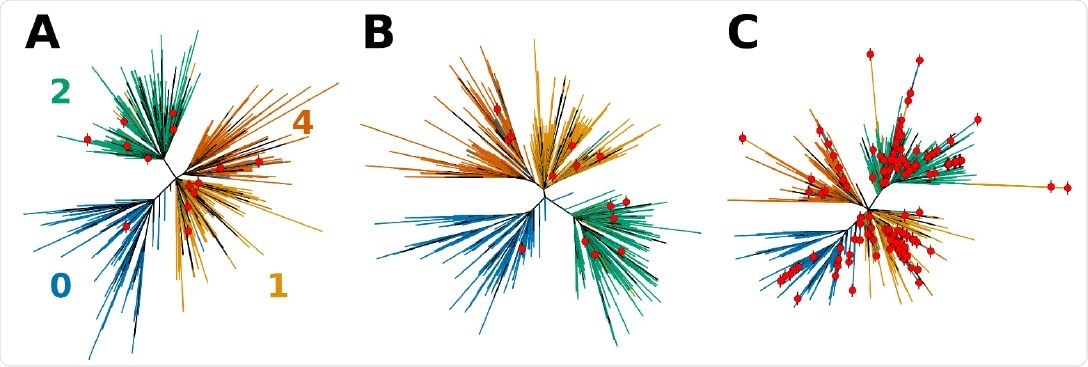
Effects of sequence composition on the topology of the phylogenetic tress for SARS-CoV-2. A tree obtained directly from NextStrain (A) is first compared to (B) the tree computed using Bolotie consensus sequences for the same set of isolates. (C) Shows a tree computed for the same set of isolates with 210 additional recombinant sequences as identified by Bolotie. Leaf nodes that correspond to recombinant genomes are labeled with red dots.
About two-thirds of the available genomes used in this study were sequenced between late March and early May. The authors believe that additional data will reveal more recombinant lineages not observed in this study. They also propose a method that can be applied to verify future SARS-CoV-2 genomes.
Using the new algorithm, Bolotie, the authors searched for such events in 87,695 complete genomes of SARS-CoV-2, from the current GISAID database. This analysis identified multiple unique cases of recombination between 4 prominent clades of the virus. They identified possibly 225 recombination events from their analysis.
Their findings show that several recombinants appear to have persisted in the population. While the cause for the demonstrated recombination could likely be homoplasies or technical artifacts, the authors show in their analysis that at least some represent actual cases of recombination. They also point out that their method used in this study is also applicable to the studies of other organisms, such as influenza. In such organisms, data is more prevalent, and parameters can be tuned with more precision.
The authors have proposed a methodology for distinguishing true recombination events from a host co-infected with several distinct lineages of a pathogen. The appearance of many events, some of which are also reported previously, are found in multiple isolates; this suggests transmission in the population. They discovered hundreds of isolates that are of a recombinant origin. They ruled out the possibility of these samples being represented co-infections.
The proposed method can be applied to verify future SARS-CoV-2 genomes before database submission.
Source:
Journal reference:
- **Rapid detection of inter-clade recombination in SARS-CoV-2 with Bolotie, Ales Varabyou, Christopher Pockrandt, Steven Lloyd Salzberg, Mihaela Pertea, bioRxiv 2020.09.21.300913; DOI: https://www.biorxiv.org/content/10.1101/2020.09.21.300913v2