The current COVID-19 pandemic has spread throughout the world. Caused by a single-stranded RNA betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is closely related to but much more infectious than the earlier highly pathogenic betacoronaviruses SARS and MERS-CoV, has impacted social, economic, and physical health to an unimaginable extent.
The virus itself has a large genome of about 30 kb, encapsidated and studded with spike glycoproteins. The spike protein exists in trimeric form, with each protomer having an S1 and S2 subunit, which bind to the angiotensin-converting enzyme (ACE) 2 receptor on the host cell surface.
The S1-ACE2 interaction occurs at the receptor-binding domain (RBD), which moves through a rigid body movement of significant amplitude in order to bind to the receptor. The RBD has both up and down positions. In the closed state, the three RBDs are down, preventing the spike protein from making any contact with the receptor. When one of the RBDs is switched into an intermediate semi-open state, its binding surface is exposed, leading to the stabilization of the RBD in the up position that favors ACE2 binding.
The ACE2 receptor is a homodimer that forms part of the cell membrane of lung, renal, heart, and intestinal epithelium. Each of the ACE2 molecules has an N-terminal peptidase domain (PD) that displays surface interaction with the spike RBD. Binding is followed by proteolytic cleavage of the S protein by the serine protease TMPRSS2, at the S1/S2 junction. This allows the S2 peptide to mediate cell membrane-virus fusion via the exposure of fusion peptides that insert into the host cell membrane.
Many vaccine development efforts are being made using computational design to bring out antigens that induce neutralizing antibodies, as well as nanoparticles, peptide inhibitors, and small molecules that counter viral attachment and entry. All these are directed at the ACE2 binding surface of the spike protein.
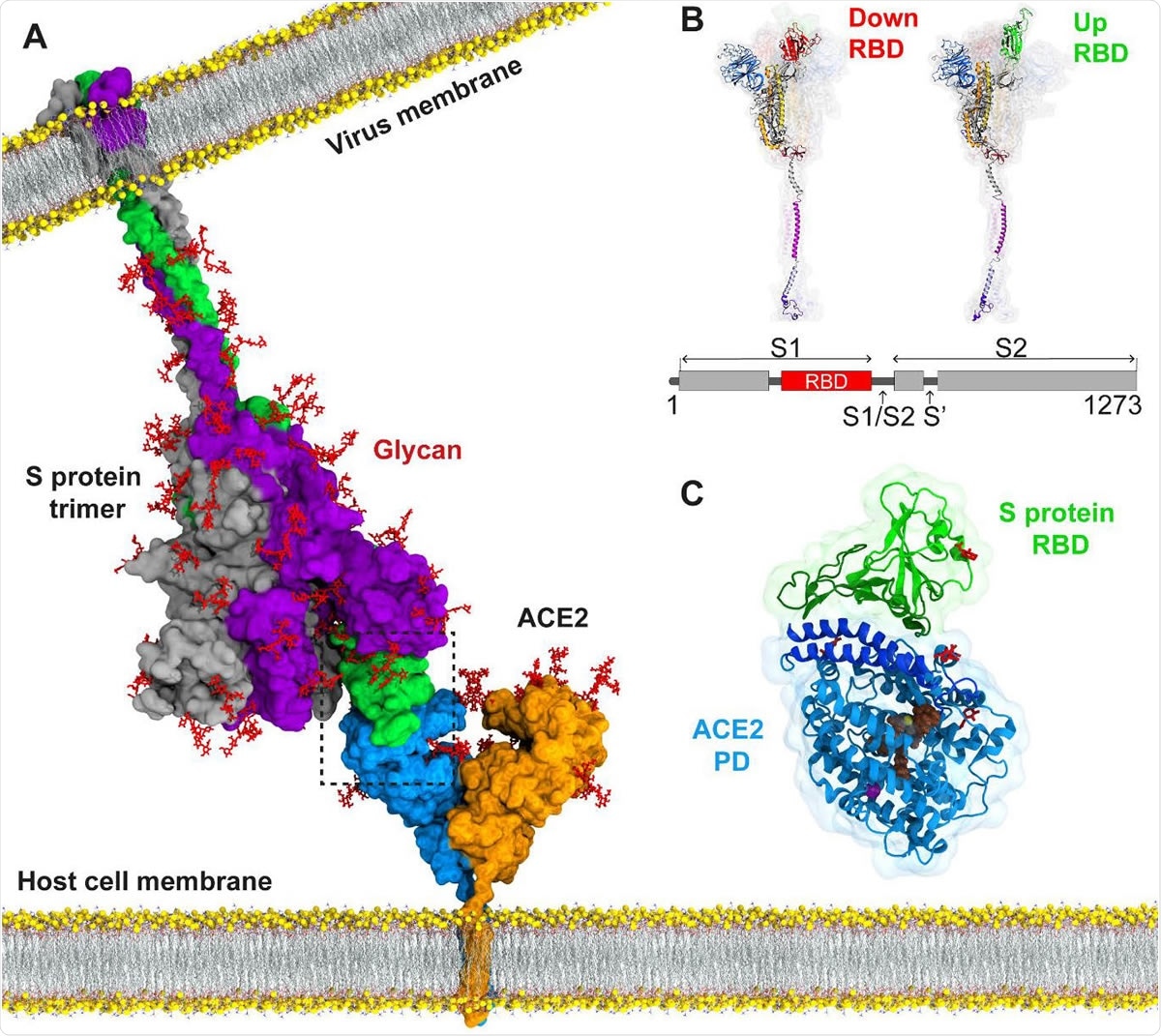
Atomic model for binding of the SARS-CoV-2 S protein to the ACE2 receptor on the host cell membrane. (A) The structure of the full-length S protein in complex with ACE2. The S protein is a homotrimer (green, purple, and grey) and embedded into the viral membrane. ACE2 is a homodimer (blue and orange) and embedded into the host cell membrane. The full length structure of the S protein in complex with ACE2 was modeled using the full length S protein model (48) and the crystal structure of the S protein RBD in complex with ACE2 (PDB ID: 6M17). Both proteins were manually inserted into the membrane by their transmembrane domains. (B) The structure of an S protomer in the down and up position of its RBD. S1/S2 and S2’ are the cleavage sites of the S protomer upon ACE2 binding. (C) MD simulations were performed for RBD of the S protein in complex with the PD of ACE2. Catalytic residues of ACE2, glycans, and Zn+2 and Cl- ions are shown in brown, red, yellow and purple, respectively.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Simulations to Understand Binding Strength
A new study by researchers at Istanbul Technical University and published on the bioRxiv* preprint server uses molecular dynamic (MD) simulations to understand the spike-ACE2 interactions in greater detail and with more accuracy. Earlier studies have used this technology to assess binding and interactions at this receptor using a homology model of the RBD-ACE2 complex. Others have used the crystal structure of this complex for MD simulations at both broad and close-up levels. This was followed up by looking at how mutations can prevent this close contact by changing the binding free energy, using further processing or bioinformatics.
However, it is not yet clear how mutations at these sites reduce the binding strength and change the pulling apart of the RBD and the ACE2. Such studies would also include an estimate of the binding free energy by measuring the work required to pull the two apart from the RBD-ACE2 complex slowly.
The researchers, therefore, carried out MD simulations for all the atoms in the complex, assisted by the fact that the RBD-PD complex structure has recently been described. They simulated the presence and absence of external forces to answer the above questions.
Interactions at the RBD-PD Complex
The researchers found a variety of interactions, including 11 hydrophobic, 8 hydrogen bonds, a couple of salt bridges, and half a dozen electrostatic interactions. The study showed some interactions not captured by the crystal structure, including 4 hydrogen, one hydrophobic, and 2 electrostatic interactions. This difference may be the result of the very different thermodynamic conditions prevailing in crystallization experiments and MD simulations.
They also clarified the effect of certain mutations on binding energies with and without external force. It became clear that the hydrophobic part of the RBD was chiefly responsible for stable RBD-ACE2 binding. Thus, this indicates a potent target for preventing SARS-CoV-2 infection.
The researchers looked at the RBD-PD complex in terms of three contact regions, CR1-3. CR2 was at the core and had much fewer interactions, with the two end regions harboring most of the interactions. CR1 exhibited predominantly hydrophobic interactions, and these are considered to be key to the RBD-PD anchoring. CR2 shows a single hydrophobic interaction, and CR3 none.
Reversing S-ACE2 Binding by Force
The researchers also simulated the unbinding of the spike protein from the PD at a uniform speed. Calculations of the average work needed, at 71 kcal/mol, shows that the S-ACE2 binding is a stable one.
Mutations at the S-ACE2 Interface Reduce Unbinding Work
The researchers examined the individual impact of each of the 16 interactions listed above using point mutations on the RBD to calculate the overall binding strength. They found in 13 of 17 mutations, the binding in the affected region and the adjacent region became looser as a probable result of disrupted interactions between the RBD and PD. The most significant impact occurred with two mutations each in the CR1 and CR3, and one in CR2. CR1 mutations also caused a marked increase in CR3 fluctuations.
The next step was to model the unbinding of the RBD in each of the variants produced by point mutations. They found a reduction of the work required to unbind the RBD from the PD by ~9% to 15% per mutation, mostly due to large fluctuations on the binding surface, underlining the vital role of hydrophobic interactions in the CR1 on complex stability.
Using double mutations, they next identified the most important interactions at the S-ACE2 interface. They discovered that the 4/7 highest mean fluctuations occurred mostly with CR1 double mutations, causing the maximum reduction in average work. Overall, these also reduced the unbinding work values, with CR1 double mutants resulting in 35% to 21% less binding energy relative to the wildtype spike-PD binding. The two salt bridges and the numerous hydrophobic interactions in CR1 are, therefore, the most important in determining the binding strength at this interface.
They also found that the S protein appears to unzip itself from the CR3 towards the CR1 in 85% of simulations, while in the rest it released itself from the CR3 last of all. An important principle here is that S-ACE2 binding is initiated by and dependent on CR1 binding. Thus, if CR1 critical residues are mutated, the binding is heavily impacted, and CR1 is detached from the PD. Alanine substitutions for the hydrophobic residues resulted in a progressive reduction of the chances that CR1 would be released last, at 55% to 65%. When certain double mutations are considered, this drops still lower, but with two other double mutations, a large CR3 fluctuation occurs, boosting the probability that CR1 will detach last. In short, both single and double mutants of the critical CR1 residues result in a large decrease in the binding free energy of this region to ACE2, indicating its anchoring role.
Both SARS-CoV-2 and SARS-CoV Spikes Show Similar Binding Strength
Some investigators have suggested that SARS-CoV-2 is more infective than SARS-CoV because its spike interacts more strongly with ACE2. The MD simulations run on RBD-PD interactions of both these viruses shows that of 15 interactions of different types, only 6 are conserved in SARS-CoV. While SARS-CoV has numerous hydrophobic interactions with ACE2, no salt bridges were observed.
Implications
The study concludes that CR1 primarily anchors the SARS-CoV-2 S protein to ACE2, and therefore the binding affinity can be markedly blocked by specific point mutations. This is supported by the observation that known nanobodies and human antibodies produce effective neutralization through its interactions with a large number of the critical residues identified in this study. It is noteworthy that some of the latter interactions are the strongest to be found in the RBD-PD complex.
The low cross-neutralization activity of SARS-CoV antibodies against SARS-CoV-2 is probably due to the lack of conservation of CR1, especially phenylalanine and glutamate. These being absent in SARS-CoV, the interface lacks corresponding hydrophobic and salt bridge interactions, though the contribution of this to higher infectivity of SARS-CoV-2 remains to be studied. Mutants in this region cannot prevent binding but may promote unbinding. Also, since there are multiple RBD-ACE2 contacts over a large surface area, blocking one region at this interface is unlikely to neutralize S-ACE2 binding.
Instead, the study concludes, “blocking of a larger surface of the CR1 region with a neutralizing antibody or nanobody is more likely to introduce steric constraints to prevent the S protein-ACE2 interactions.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Taka, E. et al. (2020). Critical Interactions Between the SARS-CoV-2 Spike Glycoprotein and the Human ACE2 Receptor. bioRxiv preprint. doi: https://doi.org/10.1101/2020.09.21.305490 https://www.biorxiv.org/content/10.1101/2020.09.21.305490v1
- Peer reviewed and published scientific report.
Taka, Elhan, Sema Z. Yilmaz, Mert Golcuk, Ceren Kilinc, Umut Aktas, Ahmet Yildiz, and Mert Gur. 2021. “Critical Interactions between the SARS-CoV-2 Spike Glycoprotein and the Human ACE2 Receptor.” The Journal of Physical Chemistry. B, May, acs.jpcb.1c02048. https://doi.org/10.1021/acs.jpcb.1c02048. https://pubs.acs.org/doi/10.1021/acs.jpcb.1c02048.