As scientists race to discover new and effective antiviral drugs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), many novel approaches are being utilized. Now, a new study published on the preprint bioRxiv* in September 2020 reports on the discovery of a potentially beneficial enzyme blocker that could inhibit SARS-CoV-2 viral replication.
Over the last ten years, covalent molecules that irreversibly inhibit viral enzymes have come into play both to understand various molecular structures and for use as therapeutics. Very often, such irreversible inhibitors mediate their effects via covalent bonds formed through a cysteine residue.
Researchers have tried to design such inhibitors and several of them have been approved by the Food and Drug Administration (FDA), including ibrutinib and afatinib. This interest is due to the fact that such drugs are sustained in their action and inhibit the activity of the enzyme on its natural substrates by competing with them for binding sites, even when the latter have high-affinity for the enzyme. They can be designed to target non-conserved cysteine residues for higher selectivity. Finally, the covalent bonds can be formed even for difficult targets such as the oncogenic K-Ras molecule with the G12C mutation.
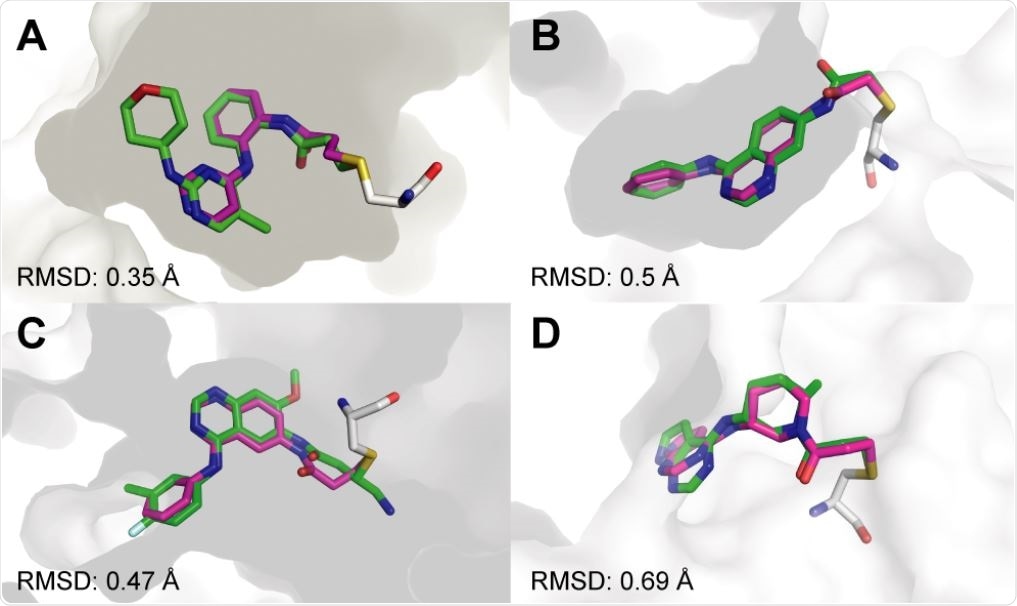
Covalentizer successfully recapitulates known covalent kinase inhibitors. Examples of covalent kinase inhibitors (green) for which covalentizer was able to find a substructure match (magenta) under the 1.5 Å threshold. A. ERK2, PDB: 4ZZO. B. EphB3, PDB: 5L6P. C. EGFR (T790M), PDB: 4I24. D. JAK3, PDB: 5TOZ. The electrophiles span acrylamides (A,D), a substituted acrylamide (C) and chloroacetamide (B).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Finding a New Covalent Inhibitor Pipeline
At present, covalent fragment libraries are being screened for covalent inhibitors, either empirically or virtually. Earlier, they were designed by adding an electrophile to an existing reversible inhibitor with a binding site adjacent to a cysteine residue.
The screening approach is laborious since the primary hits are often of low binding affinity, which means they have to be carefully adjusted to achieve adequate potency. However, if a known reversible binder is covalently derivatized, the resulting compound shows many useful properties such as time-dependent inhibition, prolonged action, better selectivity for cysteine-bearing proteins, and perhaps greater potency.
For this to be achieved, there are three aspects to be considered: the right electrophile, the best scaffold vector, and the linker, if necessary, that will provide the right positioning of the electrophile group to ensure proper binding of the scaffold with the target cysteine.
Another issue is whether the modified reversible inhibitor will be accessible for synthesis.
The Covalentizer Pipeline
To cover all these aspects, the researchers used a design algorithm that may help reveal numerous potent covalent binders to target many different molecules. This is part of a computational platform that helps identify existing reversible binders that can then be converted to covalent analogs.
This process, called covalentization, is by adding various electrophilic groups to the complex structure or the model of a ligand adjacent to a cysteine. The temporary collection of covalentized molecules is covalently docked to the target using the original structure as a reference point. This helps to identify covalent structures with a high degree of prediction confidence.
This approach was used to evaluate the whole PDB, leading to the identification of thousands of potentially irreversible inhibitors.
Four-Step Pipeline
The covalentizer pipeline consists of four steps, namely, fragmentation, electrophile diversification, covalent docking, and RMSD filtering. The ligand is thus decomposed at the bonds that are accessible for synthesis, in a repeated manner to obtain a list of substructures or vectors to which the electrophile can be added. To each list, the naked or undecorated ring system is then added, called the Murcko scaffold, to provide still more exit vectors as sources of the next additional electrophile.
The fragmentation of the ligand allows the exposure of new ligands to which the electrophile can be added, while also preventing any shift in the binding mode of the molecule relative to the original crystal structure as a result of extra constraints due to covalent bond formation. Discarding part of the original molecule may improve inhibitor potency if it prevents the steric hindrance of kinase-ligand binding when the ligand is located beyond the electrophile.
Covalent docking responds to shifts of less than an Angstrom in width. In other words, this stripped-down version may dock better than the full-length ligand, and thus this step maximizes the number of inhibitor candidates.
Diversification of the electrophiles was carried out by generating a library of hundreds of potential analogs of four kinds for each scaffold structure. These are added to the scaffold directly or via linkers.
All available cysteine rotational forms are used to prepare a structure for covalent docking, which is used to determine the final candidate covalent analogs. To increase the accuracy of covalent docking, the ligands are limited to those with 8-25 atoms and 5 or less rotatable bonds.
Finally, RMSD filtering is carried out to identify compounds that can bond covalently with the cysteine residue without a change in the original binding mode of the reversible inhibitor.
Benchmarking the Pipeline
The covalent kinase inhibitor pipeline was benchmarked by demonstrating its ability to pick out 40% of known covalent inhibitors, using their reversible component, from a 35-kinase covalent inhibitor subset of a recently reported covalent docking benchmark.
Next, they moved on to the whole PDB with almost 45,000 structures. Using the pipeline, they narrowed the search down to ~8,400 structures with one ligand atom within the sulfur atom of a free cysteine residue, binding to ~3,700 ligands.
This target collection was then clustered to eliminate redundancy, which resulted in a final set of about 2,000 structures. Several enzyme categories are included here, about 40% being transferases and 11% composed of kinases. The protocol generated 80 very high-confidence covalent analog prediction structures.
The use of longer and more diverse linkers allowed them to target more distant cysteines for a larger target space and the introduction of more diverse chemical groups. Even while restricting their application to the subset of kinases in the PDB, this step expanded the library size to several thousand per reversible inhibitor.
The total number of electrophilic compounds was thus over 3 million using di-amine linkers for the kinases. Of the final 411 protein structures that are candidates for covalent inhibition, 186 were novel, justifying this approach.
Novel Covalent Inhibitors for Various Kinases
The researchers now selected compounds with low RMSD, an intact kinase hinge binding region following electrophile addition, and synthetic ease. Synthesis and testing resulted in the demonstration of highly potent inhibitors in 5/9 predicted molecules, with IC50 in the high nanomolar to low micromolar range.
The co-crystallization of this structure validated the robust predictive property of the computational model, which can be used to develop hundreds of new inhibitors from the existing database rapidly.
A Covalent SARS-Cov-2 Main Protease Inhibitor
The reported first structure of the new SARS-CoV-2 Mpro protease was noticed to be highly similar to the active site of SARS-CoV Mpro, at a 96% sequence identity over the whole protein. Searching for a covalentized Mpro ligand, they found one prediction based on the reversible small-molecule inhibitor ML188.
Though irreversible binding was present in the covalentized form, it failed to show potent inhibition. Optimization of its reversible binding affinity produced a superior analog form with higher potency, increasing its inhibitory potential and perhaps preventing viral mutational escape in the long run. It is noteworthy that the procedure allows convenient synthesis and testing of large analog libraries systematically.
Implications
The investigators showed that automation of the protocol allows rapid coverage of the whole PDB while testing for the validity of this novel technique. The study demonstrates the predictive accuracy of the algorithm, showing through prospective testing using six actual targets that it is possible to obtain irreversible ligands without significant synthesis.
An outstanding feature is the extensive use of X-ray crystallographic structures, which generated a large library of promising predictions based on an existing reversible inhibitor, thus allowing rapid synthesis.
Both the covalentizer protocol and the database of candidate molecules are in the public domain at https://covalentizer.weizmann.ac.il. The website allows thousands of similar predictions to be seen and used by scientists the world over, as well as for newly uploaded structures to be covalentized.
Despite this early success, the protocol does not consider the synthetic feasibility of the final candidate. Again, the final compounds are typically much weaker inhibitors than the original compounds, but further optimizations are quite possible to improve the potency, such as by increased reversible recognition
The study points out the scope for the discovery of many inhibitors after the over 1.500 so far available on the website. New electrophiles and linkers can be used to change the direction of binding with the cysteine. Cysteine can even be replaced by other amino acids if new covalent ‘warheads’ are used, significantly increasing the number of ligands.
The authors sum up, “We show that using covalent docking we were able to make irreversible analogs of ligands for which a complex structure is available.”
They also discovered novel covalent kinase inhibitors, while optimizing one SARS-CoV-2 Mpro irreversible inhibitor, in a cheap, modular and rapid synthetic pathway that is. This should accelerate the discovery of covalent inhibitors targeting many other proteins.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources