The human coronavirus family consists of 7 known pathogens, for which there are no approved vaccines or specific therapeutic options. Although the seasonal human coronaviruses - OC43, HKU1, 229E, and NL63 – only cause mild respiratory infections, three highly pathogenic coronaviruses that emerged in the last two decades revealed the pandemic potential of this family of viruses.
It is known that severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) and Middle East respiratory syndrome coronavirus (MERS-CoV) can cause acute respiratory distress syndrome and even death. The fatality rates of these viruses ranged from 10 to 40%.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which caused the current COVID-19 pandemic, has a lower fatality rate but is far more transmissible than SARS-CoV-1 and MERS-CoV. So far, it has been responsible for nearly 33 million cases and 996,000 deaths worldwide.
Given the severity of the impact of these viruses on human health, we must understand the mechanism behind the invasion of host cell machinery by SARS-CoV-2 and other coronaviruses during infection. Identifying host factors common to several coronaviruses could help develop therapies that can fight current and future pandemics caused by coronaviruses.
CRISPR screens of infected cells reveal distinct viral entry factors
In a recent preprint paper published on the preprint bioRxiv,* researchers from the University of California San Francisco, Gladstone Institutes, Chan Zuckerberg Biohub, and Synthego Corporation, San Francisco, discuss how they identified some host factors common to 3 coronaviruses with the help of the gene-editing tool, CRISPR.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The team of researchers conducted parallel genome-wide CRISPR screens cells infected with SARS-CoV-2 and two seasonal common cold coronaviruses - OC43 and 229E. They were able to identify the distinct viral entry factors for all 3 viruses - ACE2 for SARS-CoV-2, glycosaminoglycans for OC43, and aminopeptidase N for 229E. Moreover, they also found that phosphatidylinositol phosphate biosynthesis and cholesterol homeostasis are critical host pathways that support infection by these three coronaviruses.
TMEM106B, the lysosomal protein, was unique to SARS-CoV-2 infection. By pharmacological inhibition of cholesterol homeostasis and phosphatidylinositol phosphate biosynthesis, the authors were able to reduce the replication of all three coronaviruses.
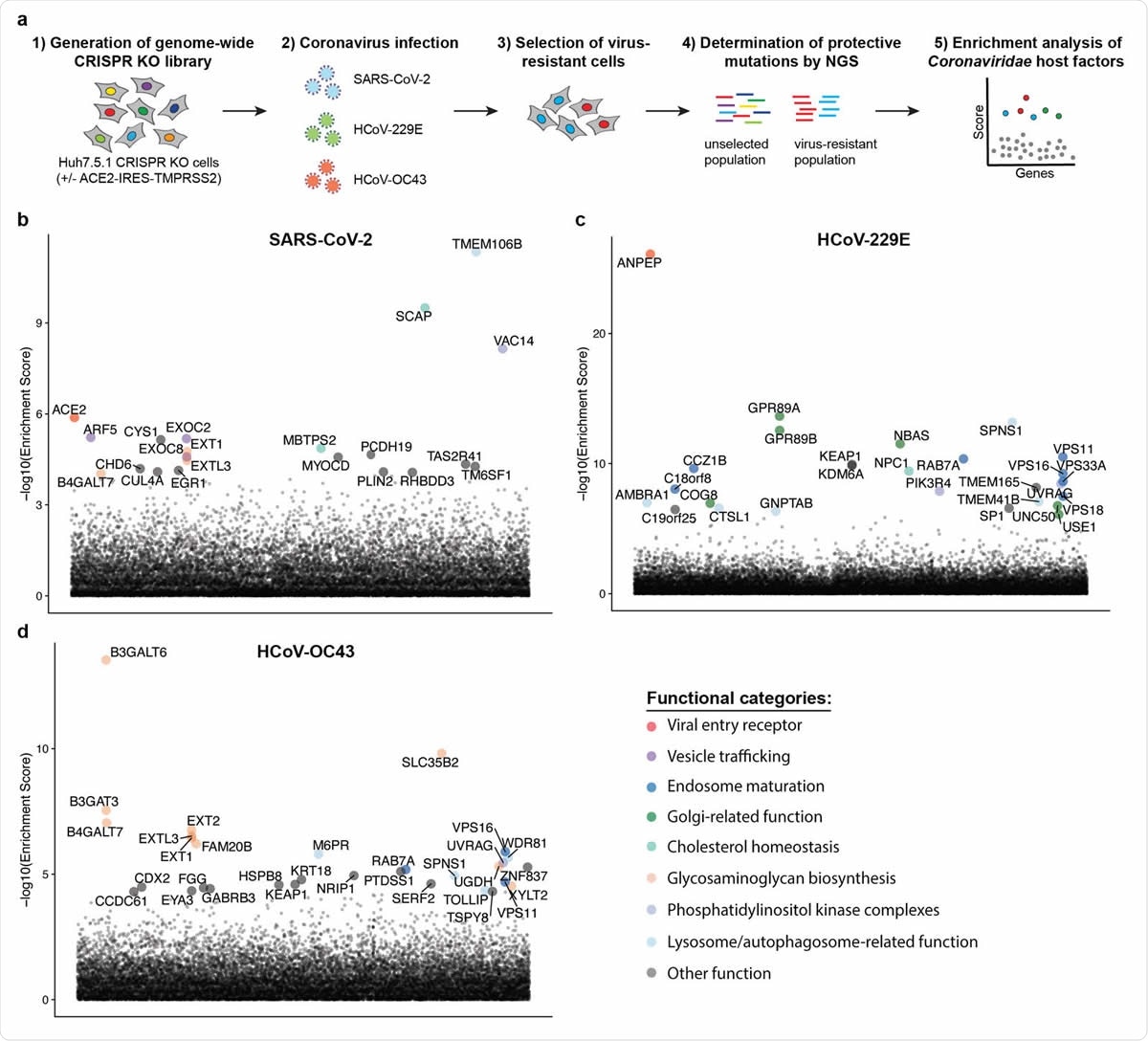
Genome-wide CRISPR KO screens in human cells identify host factors important for infection by for SARS-CoV-2, HCoV-229E and HCoV-OC43. (a) Schematic of CRISPR KO screens for the identification of coronavirus host factors. Huh7.5.1-Cas9 (with bicistronic ACE2-IRES-TMPRSS2 construct for SARS-CoV-2 and without for 229E and OC43 screen) were mutagenized using a genome-wide sgRNA library. Mutant cells were infected with each coronavirus separately and virus-resistant cells were harvested 10-14 days post infection (dpi). The abundance 347 of each sgRNA in the starting and selected population was determined by high-throughput sequencing and a gene enrichment analysis was performed. (b-d) Gene enrichment of CRISPR screens for (b) SARS-CoV-2, (c) 229E and (d) OC43 infection. Enrichment scores were determined by MaGECK analysis and genes were colored by biological function. The SARS-CoV-2 was performed once. The 229E and OC43 screens were performed twice and combined MaGECK scores are displayed.
Data suggests unique entry factors but common host pathways for 3 coronaviruses
The results of the study highlight that while the three coronaviruses depend on unique entry factors, they have a common set of host pathways that help in infection. Genes linked to cholesterol homeostasis were highlighted in all screens and the network propagation.
"Due to its involvement in multiple cellular processes including vesicular trafficking and autophagy, it remains to be determined whether coronaviruses hijack this pathway during entry or for the generation of double membrane vesicles required for the viral replication/transcription complexes."
Consistent with the findings, 2 SARS-CoV-2 interactomes revealed viral proteins binding to SCAP, which was identified as a host factor that is critical for the replication of the SARS-CoV-2 virus. The researchers argue that given the role of SCAP in viral replication, viral proteins likely positively regulate SCAP activity and cholesterol levels. These findings provide key insights into the understanding of the life cycle of coronaviruses and important directions for the development of host-directed therapies to combat coronavirus infection.
Previous studies have linked cellular cholesterol homeostasis to viral entry and membrane fusion in bunya- and hantavirus infections. This suggests a pro-viral function across different families of viruses. The team's findings were in agreement with this hypothesis. They were able to reduce infection with SARS-CoV-1 and CoV-2 spike-pseudotyped viruses by treatment with 25-hydroxycholesterol, which blocks SREBP processing and inhibits cholesterol synthesis.
The results are also consistent with that of a recent drug repurposing screen that identified more than 100 compounds, including PIKfyve inhibitors, protease inhibitors, and cholesterol homeostasis modulators, that inhibited SARS-CoV-2 replication. The functional genomics data obtained by this study suggests that the observed effects of these compounds were potentially due to inhibition of crucial host factors.
"Our results corroborate previously implicated host pathways, uncover new aspects of virus-host interaction, and identify targets for host-directed antiviral treatment."
According to the team, the study offers critical insights into host pathways, usually hijacked by coronaviruses. They identified the phosphatidylinositol PIK3C3 kinase complex as a potential therapeutic target for SARS-CoV-2 based on the 229E and OC43 screens, highlighting the value of the parallel CRISPR screening for developing novel therapies against SARS-CoV-2 and other viruses of the Coronaviridae family.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Functional genomic screens identify human host factors for SARS-CoV-2 and common cold coronaviruses Ruofan Wang, Camille R. Simoneau, Jessie Kulsuptrakul, Mehdi Bouhaddou, Katherine Travisano, Jennifer M. Hayashi, Jared Carlson-Stevermer, Jennifer Oki, Kevin Holden, Nevan J. Krogan, Melanie Ott, Andreas S. Puschnik bioRxiv 2020.09.24.312298; https://www.biorxiv.org/content/10.1101/2020.09.24.312298v1
- Peer reviewed and published scientific report.
“Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses.” 2021. Cell 184 (1): 106-119.e14. https://doi.org/10.1016/j.cell.2020.12.004. https://www.cell.com/cell/fulltext/S0092-8674(20)31626-3.