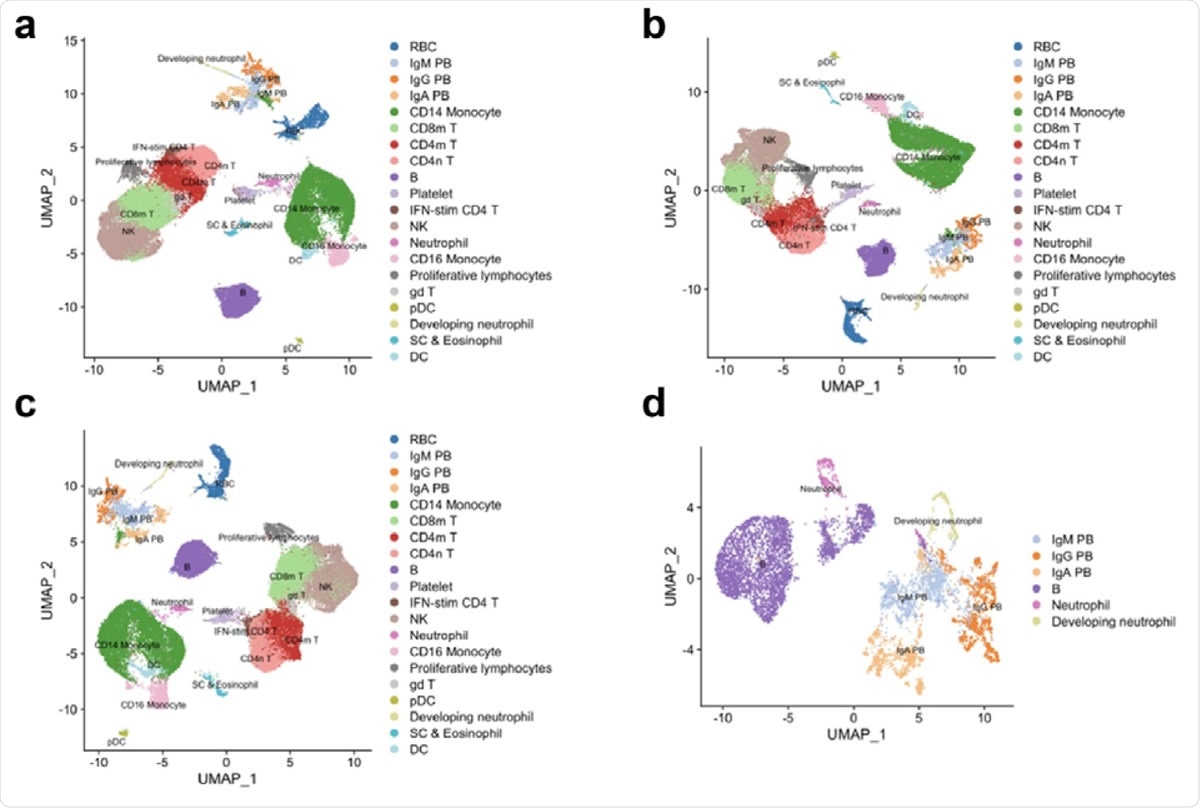
Re-analysis of scRNA-seq data from Wilk et al. show developing neutrophils do not transdifferentiate from plasmablasts. a. Reconstruction of the UMAP embedding from figure 1c of Wilk et al. b. UMAP embedding showing effect of regressing out mitochondrial genes. c. UMAP embedding showing effect of regressing out mitochondrial genes and ribosomal genes. d. UMAP embedding of neutrophils, developing neutrophils, B cells and plasma cells.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Proliferating plasma cells that are derived from lymphoid lineage B cells are called plasmablasts. In contrast, neutrophils are derived from myeloid lineage cells and are actively involved in the phagocytosis of invading pathogens. Because of the variation in cell lineages, extensive remodeling is required both at the genetic and molecular levels to convert plasmablasts into developing neutrophils. Although studies have shown that B cell progenitors and naïve B cells can be transdifferentiated into macrophages by exogenously expressing specific transcription factors, a thoroughly researched evidence is required to establish spontaneous transdifferentiation of plasma cells that are terminally differentiated.
In the current study, the scientists reanalyzed the single-cell RNA-sequencing data generated by Wilk et al. Single-cell RNA-sequencing is used to assess the transcriptional state of a wide variety of cells, and the proximity of two different cell types represents transcriptional similarities of these cells. For an accurate analysis of the data, it is vital to remove technical noise without interfering with the biological variability.
While analyzing Wilk’s data, the scientists observed that the authors have regressed out a number of unique molecular identifies (UMIs) and expressed genes from their gene expression data, in addition to regressing out ribosomal RNA and genes and mitochondrial genes. However, regressing out UMIs is not recommended, as it can lead to data overfitting.
To thoroughly reexamine the entire scenario, the current study scientists analyzed Wilk’s data by separately regressing out mitochondrial genes, regressing out mitochondrial and ribosomal genes, or without regressing out any covariates. The scientists observed that the relationship between plasmablasts and developing neutrophils no longer exists when technical noise is removed appropriately without tampering covariates.
Furthermore, Wilk’s data have shown that both developing neutrophils and plasmablasts occupy a similar manifold space, in which plasmablasts seem to be distantly associated with B cells. Moreover, the analysis performed by Wilk et al. makes a theoretical hypothesis that there is a developmental relationship between all cell types studied and that a cell lineage tree connects them. However, such a hypothesis can be made only when analyzing differentiated cells with a common ancestor in vitro. Such a hypothesis is not applicable to peripheral blood mononuclear cells as they contain several types of cells derived from different types of progenitor cells.
In the current study, the scientists further explored possible similarities between B cells, plasmablasts, neutrophils, and developing neutrophils. Surprisingly, they observed that neutrophils and B cells are not related to developing neutrophils and plasmablasts, respectively. This indicates that plasmablasts and developing neutrophils are activated in response to severe COVID-19 and possibly share the expression of a number of genes, which may have misled Wilk et al. to classify them as related cell types. Such a shared expression of genes may be an outcome of SARS-CoV-2-induced overactivation of the immune system.
Because of the high density, neutrophils cannot be generally captured in the blood preparation used to isolate peripheral blood mononuclear cells. According to the current study scientists, it is likely that developing neutrophils are similar to low-density proinflammatory neutrophils, which contain hyposegmented immature neutrophils. Therefore, it is more likely that developing neutrophils are derived from bone marrow myeloid precursors in response to severe SARS-CoV-2 infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references: