A large group of researchers from the United States reported developing an amplification-free CRISPR-based mobile phone assay for direct detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from nasal swabs. Their state-of-the-art paper is currently available on the medRxiv* preprint server.
An unrelenting coronavirus disease (COVID-19) pandemic caused by SARS-CoV-2 arose partly due to the challenges of identifying symptomatic, asymptomatic, and presymptomatic carriers of the virus – resulting in turn with their delayed isolation and massive global disease spread.
The current diagnostic gold standard is the quantitative reverse transcription-polymerase chain reaction (RT-qPCR), which is reliable and widely used for screening purposes. However, the situation in the United States shows a significant backlog, with 31% of nasal swab-based PCR tests necessitating more than four days to process.
This fact alone, coupled with a potential quick waning of the immunity after natural infection, highlights the need for rapid, point-of-care testing options that can also reliably detect SARS-CoV-2 genetic material.
In that regard, viral diagnostics may benefit from a successful bacterial strategy to destroy incoming bacteriophages (i.e., viruses that attack bacteria) and build an immunological memory, which is known as CRISPR. The latter can be exploited for the successful and quick detection of SARS-CoV-2.
To achieve the high sensitivity needed for testing purposes, current CRISPR diagnostic strategies primarily rely on pre-amplification of target ribonucleic acid (RNA) for subsequent detection by a Cas protein.
In a new and exciting study, a large research group reported the proof of concept of a rapid CRISPR-Cas13a-based diagnostic assay for the direct detection of SARS-CoV-2 RNA.
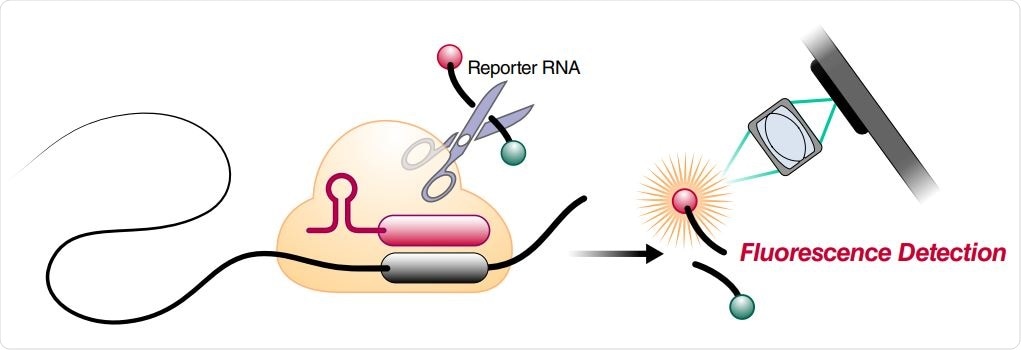
Schematic of a Cas13a (beige)-crRNA (red) RNP complex binding target RNA (black), resulting in activation of the HEPN nuclease (denoted by scissors) domain. Upon target recognition and RNP activation, Cas13a indiscriminately cleaves a quenched-fluorophore RNA reporter, allowing for fluorescence detection as a proxy for Cas13a activation and target RNA. (B) Schematic of the SARS-CoV-2 nucleocapsid (N) gene, and the corresponding locations of each crRNA spacer region.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Optimizing a compact diagnostic device
In order to achieve this goal, the researchers first needed to optimize Cas13 activation through careful selection of CRISPR RNA complexes, as well as to develop a sensitive and transportable fluorescence detection system for this novel assay.
The simplicity of the approach was demonstrated by measuring fluorescence with a mobile phone camera in a close-packed device, comprised of low-cost laser lightning and collection optics.
The researchers have also combined complexes of CRISPR RNA and Cas13 (crRNAs) that target SARS-CoV-2 RNA to enhance the sensitivity and specificity of the proposed diagnostic solutions. They have also directly quantified viral load by utilizing enzyme kinetics.
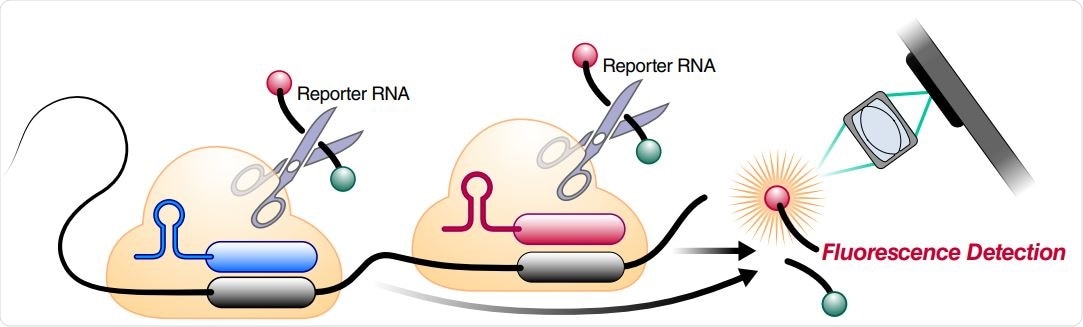
Schematic of two different RNPs binding to different locations of the same SARS-CoV-2 RNA, leading to cleavage of the RNA reporter and increased fluorescence.
Several key advances
A key advance of this approach is a successful demonstration of how these combinations of crRNAs can increase the sensitivity of Cas13a direct detection by including more Cas13a (which is a specific CRISPR-Cas13 subtype) per target RNA.
"By combining multiple crRNAs to increase Cas13a activation and analyzing the change in fluorescence over time rather than solely endpoint fluorescence, we are able to achieve SARS-CoV-2 viral RNA detection of ~100 copies/µL within 30 minutes", explained the study authors. "We also correctly identified all SARS-CoV-2 positive patient samples tested within 5 minutes", they add.
Another key advance is the possibility to translate the fluorescent signal into viral loads directly. Unlike previous diagnostic assays based on CRISPR technology, this one does not require pre-amplification of the viral genome. In other words, by directly detecting the viral RNA (without additional manipulations), we get quantitative RNA measurements instead of a simple, binary positive/negative result.
And of course, an additional important facet is that a simple mobile phone-based device can accurately read the direct-detection assay, avoiding the need for a bulky laboratory plate reader. This concept was already tried in previous molecular diagnostic approaches, primarily loop-mediated isothermal amplification.
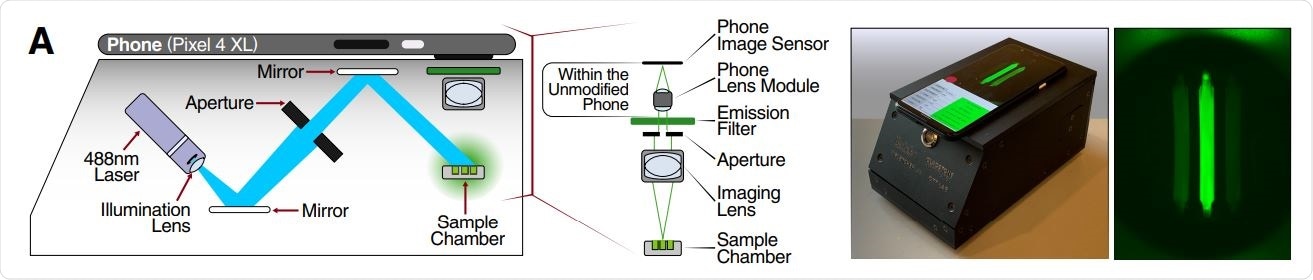
Schematic of mobile phone-based microscope for fluorescence detection showing illumination and image collection components (left). Picture of assembled device used for data collection and sample image taken by the mobile phone camera after running a Cas13a assay (right).
Fast, portable and accurate
"Here we show that direct detection of SARS-CoV-2 RNA with CRISPR-Cas13a and a mobile phone offers a promising option for rapid, point-of-care testing", summarize study authors in this promising medRxiv paper.
Mobile phone cameras became highly sensitive tools, which (together with GPS, connectivity, and data-processing abilities) have turned them into attractive devices for point-of-care disease diagnosis, especially in low-resource regions.
Combined with mobile phone-based quantification, we can really see the potential in this approach to enable fast, portable, accurate, and low-cost options for point-of-care SARS-CoV-2 screening endeavors.
Going forward, direct detection by Cas13a (as outlined in detail in this paper) may be quickly modified to target the next emerging respiratory pathogen – hopefully in time to help curtail the global spread and another potentially devastating pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Direct detection of SARS-CoV-2 using CRISPR-Cas13a and a mobile phone Parinaz Fozouni, Sungmin Son, María Díaz de León Derby, Gavin J Knott, Carley N Gray, Michael V D'Ambrosio, Chunyu Zhao, Neil A Switz, G. Renuka Kumar, Stephanie I Stephens, Daniela Boehm, Chia-Lin Tsou, Jeffrey Shu, Abdul Bhuiya, Max Armstrong, Andrew Harris, Jeannette M Osterloh, Anke Meyer-Franke, Charles Langelier, Katherine S Pollard, Emily D Crawford, Andreas S Puschnik, Maira Phelps, Amy Kistler, Joseph L DeRisi, Jennifer A Doudna, Daniel A Fletcher, Melanie Ott medRxiv 2020.09.28.20201947; doi: https://doi.org/10.1101/2020.09.28.20201947
- Peer reviewed and published scientific report.
Fozouni, Parinaz, Sungmin Son, María Díaz de León Derby, Gavin J. Knott, Carley N. Gray, Michael V. D’Ambrosio, Chunyu Zhao, et al. 2020. “Amplification-Free Detection of SARS-CoV-2 with CRISPR-Cas13a and Mobile Phone Microscopy.” Cell, December. https://doi.org/10.1016/j.cell.2020.12.001. https://www.cell.com/cell/fulltext/S0092-8674(20)31623-8.