A team of scientists of the University of Chicago and the University of California San Diego has developed a coarse-grained (CG) model of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virion. The model can be used as a valuable resource to perform multiscale simulations of the SARS-CoV-2 virion. The study is currently available on the bioRxiv* preprint server.
SARS-CoV-2, the causative pathogen of the coronavirus disease 2019 (COVID-19) pandemic, is a positive-sense single-stranded RNA virus with a genome size of 30 kb. To obtain a detailed overview of SARS-CoV-2 viral proteins and their functions, many structural biology studies have been performed using cryo-electron microscopy and x-ray crystallographic techniques. Simultaneously, many computational biology studies have also been done to predict unresolved genome regions using multiple protein folding algorithms. However, given the difficulty in simulating biomolecules at atomic resolutions, all-atom molecular dynamic simulation of various viral processes, such as virion assembly, budding, entry, and fusion, is fundamentally a challenging task.
To overcome these challenges, the current study aimed at building a bottom-up CG model of the SARS-CoV-2 virion using publicly available experimental structural data and atomic simulation data about key viral structural proteins, including the spike, membrane, nucleocapsid, and envelop proteins. In the current CG model of the SARS-CoV-2 virion, the scientists derived molecular interactions between CG particles using phenomenological, experimental, and atomistic simulation approaches.
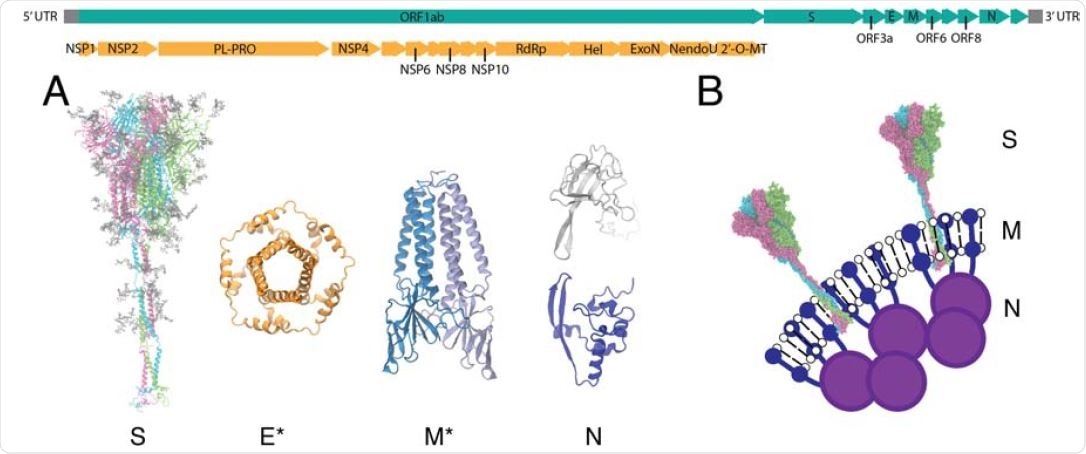
Viral proteins of SARS-CoV-2. The genome of SARS-CoV-2 is shown in the top panel. Nonstructural proteins (NSPs) encoded in the open reading frame (ORF) 1ab are colored in orange, and the full genome is in teal. (A) All-atom models of the structural proteins of SARSCoV- 2 consisting of the S, E, M and N proteins. Asterisks indicate homology modeled protein structures for E and M (34). (B) Schematic of the virion surface from cryo-EM images of the virion, adapted from Ref. (19).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Current study design
Initially, the scientists developed all-atom models of SARS-CoV-2 structural proteins using a combination of cryo-electron microscopy, x-ray crystallography, and computational simulations. Afterward, the CG models were developed by simulating and coarse-graining the all-atom protein models.
Benefits of CG simulations
Complex biological systems, such as virions, are difficult to capture by all-atom simulations as well as experimental methods because of a wide range of physical and chemical transitions involved. The same is true for complex biological processes that involve multiple conformational changes. For example, the proteolytic cleavage of the SARS-CoV-2 spike protein results in a large conformational change, which is essential for the interaction between the viral spike protein and host cell angiotensin-converting enzyme 2 (ACE2), as well as the insertion of the fusion peptide into the host cell membrane. However, such large-scale biological events are difficult to be captured through all-atom simulations might be because of longer timescales or free energy barriers.
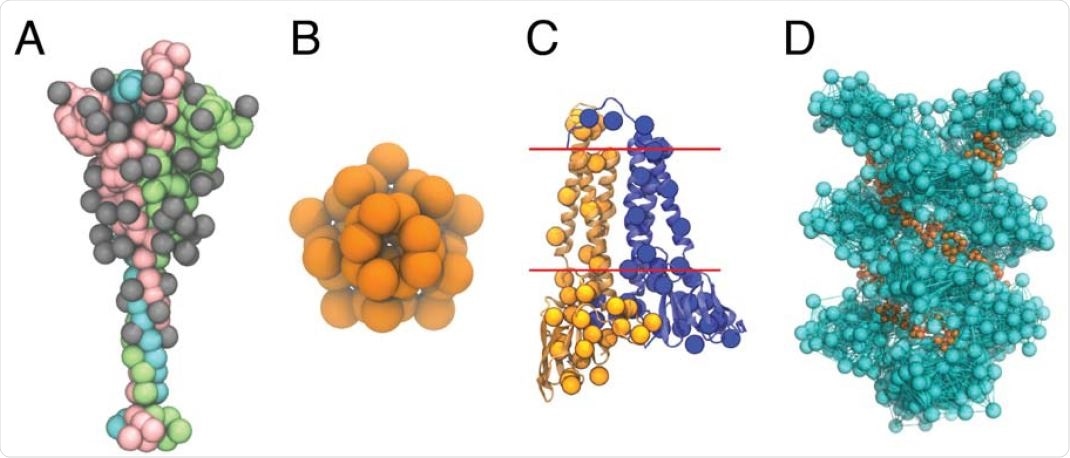
CG models of the SARS-CoV-2 structural proteins. (A) The CG model of the S protein trimer in the open state. The protein monomers are depicted as pink, green, and cyan beads, respectively; the monomer in pink has an exposed receptor binding domain. Each of the 22(x3) N-linked glycans are depicted as grey beads. (B) The CG model of the pentameric E protein is depicted as orange beads. (C) The CG M dimer model is depicted as yellow and blue spheres, overlaid on top of the AA model of the M dimer. Each monomer has 36 CG sites, and the red lines indicate the approximate positions of the transmembrane region. (D) The CG model of the N protein CTD helix in complex with viral RNA. The N protein helix and bonds derived from the hENM are depicted in cyan, while the RNA is depicted as orange beads.
In contrast to all-atom simulations, CG methods allow simulating complex biological processes at various granularity levels. CG models can be developed from all-atom models (bottom-up) by reducing the dimensionality of all-atom models into a reduced number of pseudo-atoms and averaging out fast local motions.
Important observations
In the current CG model of the SARS-CoV-2 virion, a particle-based phenomenological simulation was used to describe the lipid envelop, and membrane and envelop proteins were modeled as rigid bodies. Moreover, relative entropy minimization methods based on microsecond all-atom spike protein simulations were used to describe intra-spike interactions.
An All-atom simulation of the helical oligomer-RNA complex was used to develop the nucleocapsid protein model. Attractive Gaussian potentials between lipid tails and transmembrane protein domains were used to simulate the interaction between lipids and structural proteins.
The CG model developed in the current study is a preliminary form of the virion, which can be advanced over time with the availability of new all-atom simulation data and experimental structural data of various SARS-CoV-2 proteins.
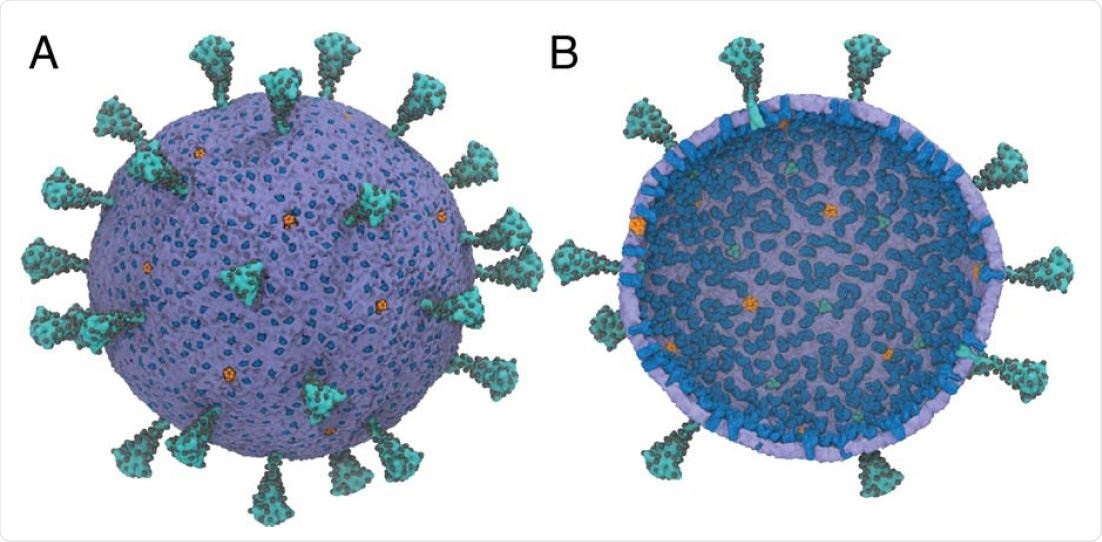
A multiscale model of the SARS-CoV-2 virion. (A) Exterior view of the SARS-CoV-2 virion. (B) Interior view of the SARS-CoV-2 virion. Spike (S) protein trimers are depicted in teal with the glycosylation sites represented as black spheres. Membrane (M) protein dimers are in blue, with pentameric envelope (E) ion channels in orange. The density of S,M, and E proteins was chosen to be consistent experiments (38–40). N proteins are not shown. The diameter of the membrane envelope is approximately 100 nm and 120 nm including the S proteins on the virion surface.
Study significance
CG models of various viral proteins can provide essential information about a wide variety of viral mechanisms, deciphered experimentally to develop therapeutic interventions. For example, CG models of the human immunodeficiency virus (HIV) have been used previously to understand the capsid self-assembly and to identify inhibitors of the viral activity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources