Researchers in the United States have made an important discovery about the processing and presentation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral peptides that provides a better understanding of the immune response to coronavirus disease 2019 (COVID-19) and may help facilitate the design of improved vaccines.
The team has presented the first detailed analysis of viral peptides presented by class I human leukocyte antigen (HLA-I) on human cells infected with SARS-CoV-2.
Interestingly, the researchers found that nine of the viral peptides are derived from non-canonical open reading frames (ORFs) in the viral Spike protein and Nucleoprotein. Spike is the surface structure SARS-CoV-2 uses to bind to and infect host cells, while Nucleoprotein is involved in packaging viral RNA and the release of virions.
The team says the finding suggests that studies of the T cell response among COVID-19 patients that currently only focus on canonical viral ORFs exclude a significant number of virus-derived HLA-I epitopes.
The current pools of SARS-CoV-2 derived HLA-I peptides that enable T-cell activating viral signatures to be identified may be incomplete, say Pardis Sabeti (Broad Institute of MIT and Harvard, Cambridge) and colleagues.
New approaches that directly identify naturally presented SARS-CoV-2 peptides would help to inform the design of future pools to provide a more detailed characterization of T cell responses in COVID-19 patients. Such approaches would also facilitate the design of more effective vaccines, say the researchers.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
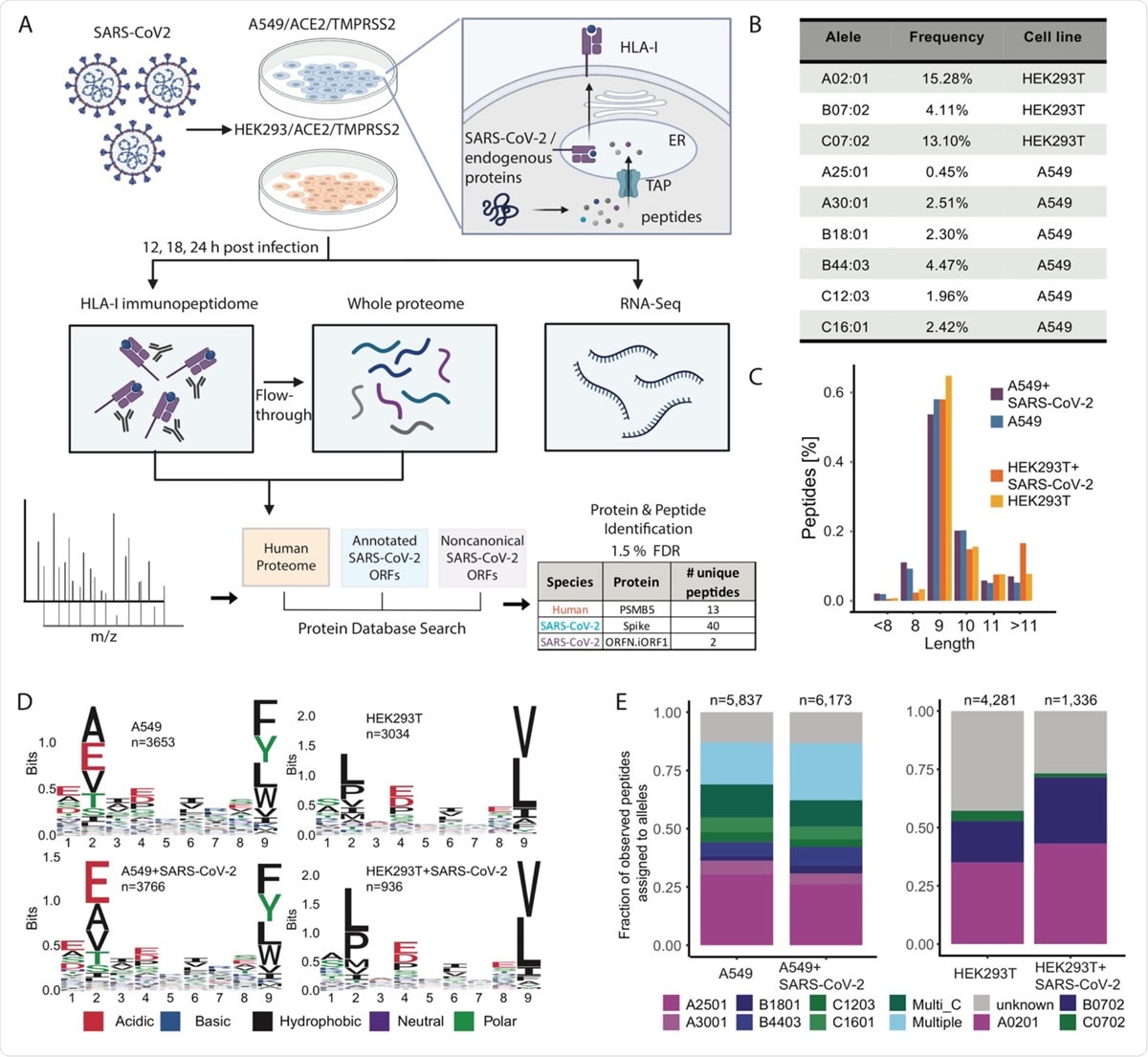
Experimental design and measurements of HLA-I immunopeptidome, whole proteome and RNA-seq in SARS-CoV-2 infected cells. (A) Schematic representation of the experiment and the antigen presentation pathway. A549/ACE2/TMPRSS2 and HEK293T/ACE2/TMPRSS2 cells were infected with SARS-CoV-2 (Washington strain, accession number MN985325) at Multiplicity of Infection (MOI) of 3 in a BSL3 facility and harvested at 12, 18 and 24hpi (hours post infection). For HLA-I immunopeptidome measurements cells were lysed and HLA-I peptide complexes were immunoprecipitated. The flow-through after immunoprecipitation was collected for whole proteome measurements. For RNA-seq we added Trizol to infected cells, purified the RNA and performed strandspecific short reads sequencing. MS/MS spectra of immunopeptidome and proteome analysis were searched against a protein database including the human proteome, canonical and non-canonical SARS-CoV-2 ORFs and filtered at 1.5% FDR. (B) Population frequency of the 9 endogenous HLA-I alleles expressed in A549 and HEK293T cells. (C) Length distribution of HLA peptides in infected and naive cells. (D) Motif of 9-mer sequences identified in infected and naive cells. (E) Fraction of observed peptides assigned to alleles using HLAthena prediction (percentile rank cutoff <0.5) for immunopeptidome of infected and uninfected cells.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The limitations of current approaches
As research efforts to develop effective vaccines for SARS-CoV-2 continue, it is essential to understand the role that T cells play in establishing long-term immunity against the virus.
Following infection, viral proteins are processed and presented on the host cell surface by HLA-I for recognition by cytotoxic T cells, which then induce an immune response to combat the virus.
To date, most studies that have examined the interaction between T cells and SARS-CoV-2 antigens have used bioinformatic predictions of HLA-I binding affinity. However, this approach is limited because it does not account for all of the steps involved in antigen processing and presentation, and the average positive predictive values achieved across HLA alleles are still only about 64%.
These prediction models also do not account for how viruses might alter cellular processes and affect antigen presentation.
“For example, viruses can attenuate translation of host proteins, downregulate the proteasome machinery, and interfere with HLA-I expression,” say Sabeti and team. ”These changes shape the collection of viral and human-derived HLA-I peptides presented to the immune system.”
Experimental measurements of the SARS-CoV-2 peptides presented on host cells are therefore, essential to understanding how the immune system responds to infection.
Mass spectrometry-based HLA-I immunopeptidomics is a direct and unbiased approach to discovering endogenously presented peptides and to establishing the processes that govern antigen processing and presentation.
“Leveraging HLA-I immunopeptidome datasets to learn virus-specific antigen processing rules will improve our ability to predict viral epitopes accurately and utilize them to study immune responses in COVID-19 patients,” write the researchers.
The analysis of mass spectrometry-derived data requires selecting a set of viral ORFs to examine, and until now, such analyses have primarily been focused on canonical ORFs.
However, over the last ten years, genome-wide sequencing studies have revealed a surprising number of non-canonical ORFs in viral genomes, the function of which remains largely unknown.
What did the current study find?
Sabeti and colleagues have now presented the first detailed analysis of HLA-I immunopeptidome in two SARS-CoV-2-infected human cell lines using mass spectrometry, RNA-sequencing, and global proteomics measurements.
The team identified viral HLA-I peptides derived not only from canonical ORFs but also from non-canonical ORFs in SARS-CoV-2 Spike and Nucleoprotein that are not captured by current vaccines.
“Remarkably, 9 of 29 viral peptides detected are derived from internal out-of-frame ORFs in S [Spike] (S.iORF1/2) and N [nucleoprotein] (ORF9b),” write the researchers.
The team says this finding implies that current studies investigating T cell responses in COVID-19 patients that focus only on the canonical viral ORFs exclude a significant number of virus-derived HLA-I epitopes.
The researchers estimated that based on the list of endogenously processed and presented SARS-CoV-2 peptides they identified, a pool of 24 peptides would provide one or more peptides for at least one HLA-I allele in 99% of the human population.
What are the implications of the study?
Sabeti and colleagues say the study findings could be used to facilitate the data-driven selection of peptides for immune monitoring and to improve the design of effective vaccines.
“These findings hint that current peptide pools may be incomplete,” write the researchers.
“Therefore, unbiased approaches such as LC-MS/MS immunopeptidomics that directly identify naturally presented SARS-CoV-2 peptides on host cells can inform the design of future pools to allow more comprehensive characterization of T cell responses in COVID-19 patients", concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sabeti P, et al. SARS-CoV-2 infected cells present HLA-I peptides from canonical and out-of-frame ORFs. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.10.02.324145
- Peer reviewed and published scientific report.
Weingarten-Gabbay, Shira, Susan Klaeger, Siranush Sarkizova, Leah R. Pearlman, Da-Yuan Chen, Kathleen M. E. Gallagher, Matthew R. Bauer, et al. 2021. “Profiling SARS-CoV-2 HLA-I Peptidome Reveals T Cell Epitopes from Out-of-Frame ORFs.” Cell 184 (15): 3962-3980.e17. https://doi.org/10.1016/j.cell.2021.05.046. https://www.cell.com/cell/fulltext/S0092-8674(21)00701-7.