The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is predicted by some experts to be around for several years. This alarming hypothesis compels rapid and successful therapeutic interventions to mitigate this infection. In this effort, various recombinant monoclonal antibodies of human origin that neutralize SARS-CoV-2 infection have been isolated from convalescent patients. However, the need for dedicated monoclonal antibodies in molecular pathology research is not fully addressed; these antibodies can be used to dissect the molecular mechanism of the virus life cycle.
This technology provides a reliable source of mouse monoclonal antibodies (Kohler & Milstein, 1975). The humanized mouse antibodies and subsequent generation of fully human antibodies by various techniques are widely used for various therapies. For example, Palivizumab, a humanized mouse monoclonal antibody neutralizing respiratory syncytial virus (RSV), is used in clinical settings prophylactically to protect vulnerable infants. With this approach, passive immunization with a monoclonal antibody is currently considered as a treatment for COVID-19.
While a large number of neutralizing antibodies are reported to inhibit SARS-CoV-2 infection, the authors argue that there is still an overwhelming lack of data on a well-characterized antibody available for basic research techniques such as western blotting, immunofluorescence, and immunoprecipitation to study the viral life cycle. With this need in mind, they establish six monoclonal antibodies against the spike glycoprotein of SARS-CoV-2 and report the attenuation of viral interaction with cells in vitro. The results observed in the study are summarized below.
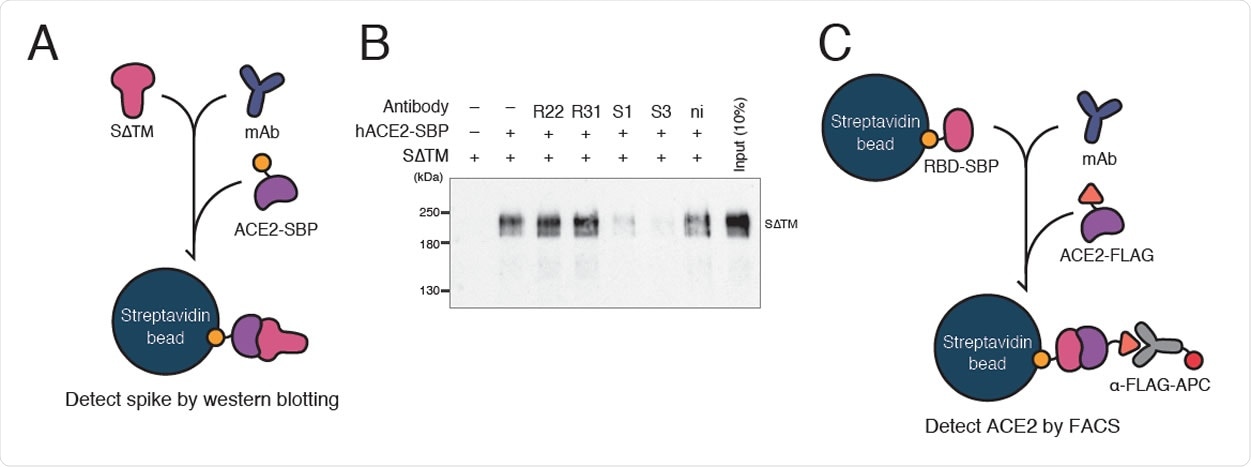
Inhibition of ACE2-spike interaction by S1D7 and S3D8 A. A schematic of the spike pull-down assay designed to evaluate inhibition of ACE2- spike binding by monoclonal antibody. Spike glycoprotein lacking TM domain (SΔTM) was mixed with a monoclonal antibody. ACE2-SBP was applied to capture SΔTM onto streptavidin beads competitively. Captured SΔTM was detected by WB as a measurement of the antibody’s inhibitory ability. S1, S1D7; S3, S3D8; ni, non immune mouse IgG. B. WB of spike pull-down assay using antibody R52. In the presence of clones S1D7 and S3D8, ACE2 was not able to pull down SΔTM. C. Schematic of bead-based neutralization assay designed to quantify inhibition of ACE2-RBD binding by monoclonal antibody. RBD-SBP glycoprotein immobilized on streptavidin beads was mixed with a monoclonal antibody. ACE2-FLAG was applied to bind competitively with RBD. ACE2-RBD binding was quantified by measuring the signal given by an anti-FLAG antibody conjugated with APC fluorophore using FACS.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Production of six monoclonal antibodies against spike glycoprotein
The SARS-CoV-2 spike glycoprotein is a homotrimeric fusion protein composed of two subunits: S1 and S2. The researchers followed the methodology developed by Wrapp et al., 2020 - in which the SARS-CoV-2 spike protein was engineered to form a stable homotrimer that was resistant to proteolysis during protein preparation. The mice were immunized with these recombinant spike proteins to generate anti-SARS-CoV-2 antibodies, followed by cell fusion to generate a hybridoma-producing antibody. In this study, after the culture supernatants were pre-screened, six monoclonal hybridomas were isolated and evaluated.
The antibodies were purified from culture supernatant and characterized in detail, with ELISA and WB performance.
S1D7 and S3D8 antibodies showed higher performance on IP (immunoprecipitation) and IF (immunofluorescence)
To understand the molecular mechanism of SARS-CoV-2 infection, especially the cell entry, where these proteins play a significant role, the antibody must recognize the intact tertiary structure of spike proteins. The IP activity of antibodies can be correlated with capturing the native structure of the target protein and utilizing the infection. All clones were capable of immunoprecipitation with the RBD and ectodomain (S∆TM) glycoproteins with a differential IP efficiency. The immunofluorescence assay reflects the cellular localization of spike proteins, which elucidate the mechanism of packaging and maturation of variance during release from the cellular membrane.
ACE2-Spike binding inhibition of the monoclonal antibodies
The authors show in this study that the spike-ACE2 interaction is intercepted by competitive binding between neutralizing antibodies and spike glycoprotein - thus, inhibiting the spike-ACE2 binding or even neutralizing SARS-CoV-2 infection. They also quantified the inhibition ability, using a bead-based neutralization assay by measuring the amount of ACE2 bound to RBD beads after blocking with monoclonal antibodies.
S1D7 and S3D8 showed neutralizing activity against SARS-CoV-2
Then, the researchers tested for the antibody's ability to inhibit SARS-CoV-2 infection in VeroE6/TM2 cells. These cells are more prone to virus infection than the commonly adopted VeroE6 cell line. Two of the six antibodies used in the study blocked SARS-CoV-2 infection significantly with IC50 values of 405.2 ng/mL and 139 ng/mL, respectively. When the researchers mixed these two antibodies, the cocktail showed intermediate neutralizing activity, suggesting a shared inhibitory mechanism.
The authors state that the mouse antibodies used in this study do not apply to clinical treatment. However, they can be used for investigating the mechanism of immune responses during passive immunization using mouse models for SARS-CoV-2 infection. An in-depth look into the fundamentals of how this virus attacks has significant implications for developing countermeasures and also diagnosis, vaccine design, and drug discovery.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Potent mouse monoclonal antibodies that block SARS-CoV-2 infection, Youjia Guo, Atsushi Kawaguchi, Masaru Takeshita, Takeshi Sekiya, Mikako Hirohama, Akio Yamashita, the Keio Donner Project, Haruhiko Siomi, Kensaku Murano, bioRxiv 2020.10.01.323220; doi: https://doi.org/10.1101/2020.10.01.323220
- Peer reviewed and published scientific report.
Guo, Youjia, Atsushi Kawaguchi, Masaru Takeshita, Takeshi Sekiya, Mikako Hirohama, Akio Yamashita, Haruhiko Siomi, and Kensaku Murano. 2021. “Potent Mouse Monoclonal Antibodies That Block SARS-CoV-2 Infection.” Journal of Biological Chemistry 296 (January): 100346. https://doi.org/10.1016/j.jbc.2021.100346. https://www.jbc.org/article/S0021-9258(21)00118-6/fulltext.