Since its emergence in December 2019, highly infectious and deadly SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has infected more than 36 million people and claimed more than 1 million lives worldwide. Although it belongs to the same viral genus, SARS-CoV-2 differs significantly from other coronaviruses, such as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), in terms of transmissibility and infectivity.
Overall, the genome of SARS-CoV-2 encodes 25 non-structural proteins and 4 structural proteins, including the spike, membrane, nucleocapsid, and envelop proteins. Besides spike protein, the nucleocapsid is the most abundant immunogenic protein that encapsulates the viral RNA. Nucleocapsid protein plays many essential roles, including viral replication/proliferation, virion assembly, host cell cycle regulation, and host immune system evasion. Interestingly, studies have found that host adaptive immune responses to nucleocapsid is more sensitive than the spike protein, which makes nucleocapsid an important therapeutic target. Regarding other viruses, many in vitro studies have shown that molecules targeting nucleocapsid proteins are effective in inhibiting viral replication, transcription, and virion assembly.
Given the significance of nucleocapsid protein in anti-viral drug development, the current study aimed at deciphering the involvement of nucleocapsid protein in the SARS-CoV-2 lifecycle.
Important observations
There are two major structural domains of the nucleocapsid protein: the RNA binding domain and the oligomerization domain, separated by an extended, flexible linker region. The RNA binding domain interacts with both host and viral RNA, whereas the oligomerization domain forms the viral core.
Using native mass spectrometry, the scientists observed that the dimers of full-length nucleocapsid protein interact with RNA, indicating that dimers, and not monomers, are the functional unit of ribonucleoprotein assembly. Moreover, they observed that nucleocapsid protein dimers preferentially bind RNA through the GGG motif, which is known to form stem-loop structures.
Interestingly, they observed that autoproteolytic events occur within the linker region of full-length nucleocapsid protein, generating at least five proteoforms. To identify proteolytic cleavage sites, they next sequenced these proteoforms using electron transfer dissociation and higher-energy collision-induced dissociation and validated the sequences by peptide mapping. They observed that proteolytic cleavage sites are highly conserved in the SARS-CoV-2 genome. In general, the cleavage sites were found to be immediately next to a hydrophobic residue.
Proteoforms of nucleocapsid protein
Of five proteoforms (N1-209, N1-220, N1-273, N156-419, and N1-392), N1-209 and N1-220 mainly contain the RNA binding domain; N1-273 contains a significant part of the RNA binding domain, followed by the linker region and a small part of the oligomerization domain; and N156-419 primarily contains the oligomerization domain and a small part of the RNA binding domain.
Regarding functional properties, the scientists observed that alike full-length nucleocapsid protein, the N-terminal proteoforms preferentially interact with RNA via the GGG motif and do not undergo pH-dependent alteration in the oligomeric state. In contrast, the oligomeric state of the C-terminal proteoforms was found to be sensitive to both high and low pH conditions.
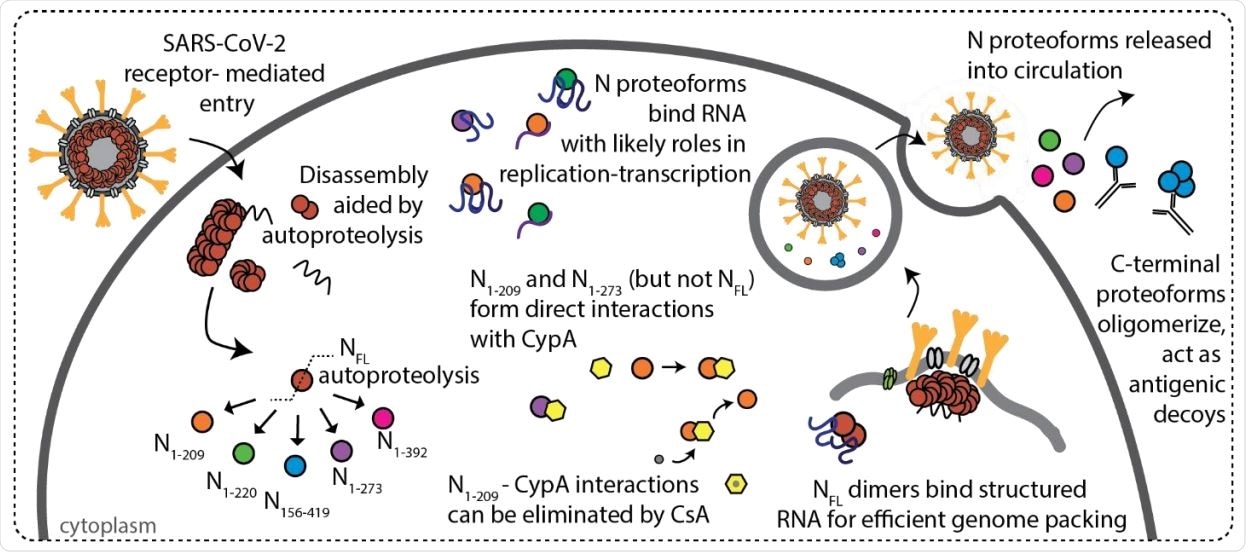
Scheme depicting features of SARS-CoV-2 N protein during infection. N protein undergoes autoproteolysis in a highly specific fashion to produce N1-209, N1-220, N1-273, N156-419, and N1-392. NFL and N proteoforms bind RNA with a preference for structured RNA and NFL dimers are likely functional units of assembly in ribonucleoprotein complexes. Immunophilin CypA binds directly to N1-209 and N1-273, but not NFL or N1-220, and the interaction can be inhibited through addition of cyclosporin A (CsA). N156-419 is readily detectable by antibodies, suggesting a role in immune evasion via the formation of antigenic decoys.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Immunogenicity and antigenicity
Using mass spectrometry, the scientists observed that although full-length nucleocapsid protein binds to an anti-nucleocapsid monoclonal antibody, proteoforms lack C-terminal residues do not interact with the antibody. This indicates that the antigenic region of nucleocapsid protein is located toward the C-terminus.
Regarding the interaction between proteoforms and vital replication components of SARS-CoV-2, such as cyclophilin A, the scientists observed N-terminal proteoforms (N1-209 and N1-273) directly interact with cyclophilin A, and this interaction can be inhibited by administering immunosuppressants.
Study significance
Overall, the current study findings indicate that N-terminal proteolytic products generated by autoproteolysis of SARS-CoV-2 nucleocapsid protein can evade the antibody-mediated immune responses by removing the C-terminal residues. Also, these N-terminal proteolytic products can facilitate viral proliferation by directly binding vital components of the viral replication process.
The study also emphasizes that inhibition of the proteolytic process can suppress the immune evasion and proliferation of SARS-CoV-2, it can be ventured as a new therapeutic intervention.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources