Researchers in the United States has provided a detailed catalog of the interactions that occur between host cell proteins and RNA and the RNA of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during the course of infection.
SARS-CoV-2 is the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic that is increasingly posing a threat to global public health and the worldwide economy.
By integrating the analysis across various time points, species, and other positive single-stranded RNA viruses, the researchers have identified both shared and SARS-CoV-2-specific patterns of RNA–host protein interactions.
The team, from Stanford University and Yale University, says the findings may help to inform future studies aiming to understand viral pathogenesis and potential therapeutic strategies to combating SARS-CoV-2 infection.
A pre-print version of the paper is available in the server bioRxiv*, while the article undergoes peer review.
Researchers are urgently trying to understand host cell infection and antiviral immunity
As the COVID-19 pandemic continues to sweep the globe with no vaccine or effective treatment yet in sight, researchers are urgently trying to understand the molecular mechanisms required for host cell infection and antiviral immunity.
Although the genomes of positive single-stranded RNA viruses share similar replication strategies, there is remarkable variation in the health outcomes these pathogens cause.
For instance, mosquito-borne flaviviruses such as Dengue and Zika cause systemic disease, while human coronaviruses such as SARS-CoV-2 generally cause respiratory symptoms.
The infection process is complex and often highly specific to the individual virus.
After a virus binds to and enters a host cell, the viral genome remodels host cellular pathways in order to express, replicate, and produce new virions.
Once viral RNA transcripts are deposited in host cells, they eventually produce viral protein products.
“Together, these RNA and protein species remodel the cell to facilitate the viral life cycle,” write Ryan Flynn (Stanford University) and colleagues.
Some studies have recently described the viral products encoded by SARS-CoV-2 RNA and their interactions with host partners, but the precise interactions of SARS-CoV-2 viral RNA (vRNA) with host partners are not well understood.
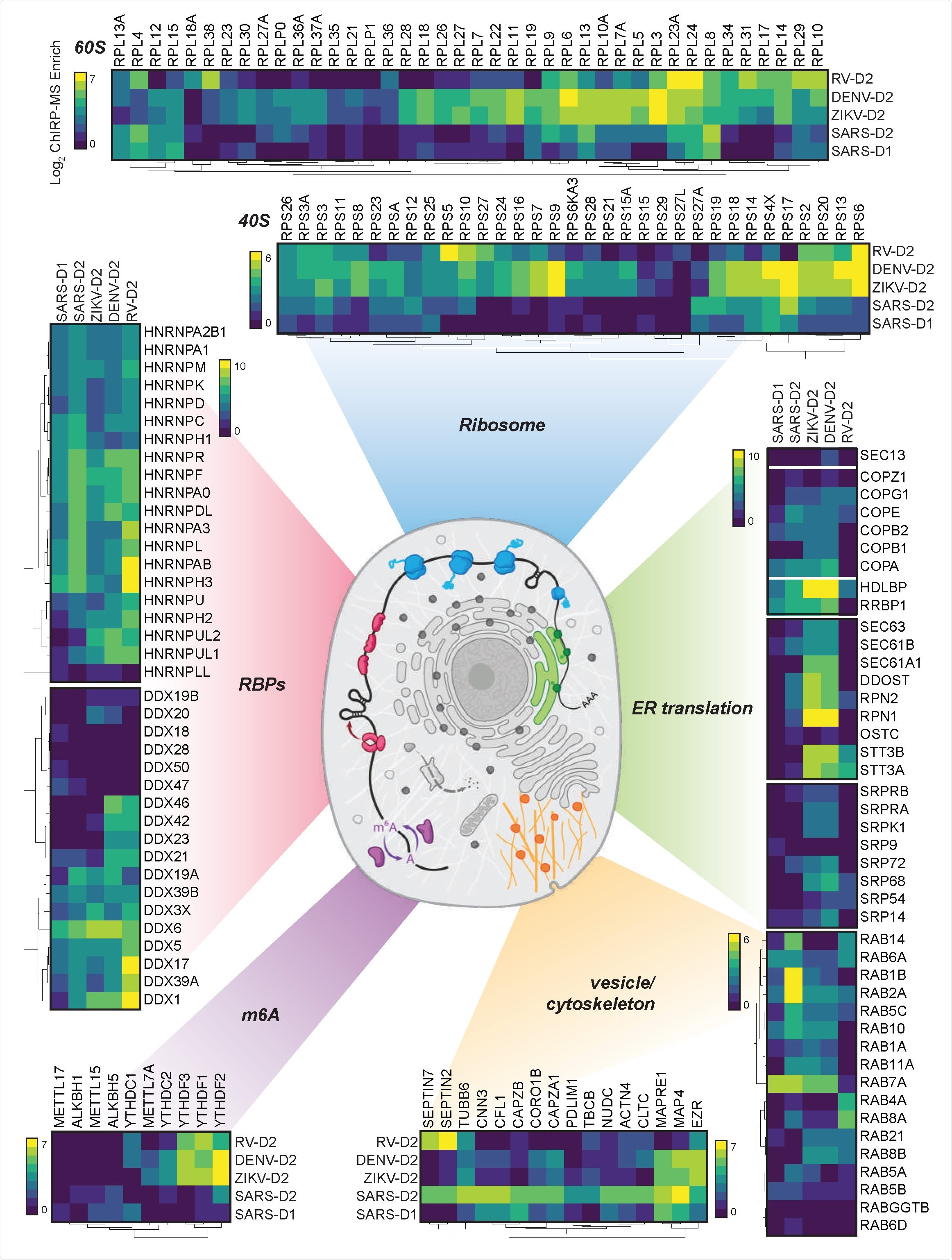
Cellular context of expanded interactomes across viruses. Selected groups of proteins, their enrichment in SARS-CoV-2, Zika, Dengue, and Rhinovirus ChIRP, and their approximate subcellular localization. Heat map colors indicate the log 2 ChIRP-MS enrichment values. Each heatmap has a separate scale bar.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
What did the current study involve?
Now, Flynn and colleagues have used comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-ms) to define both the shared and SARS-CoV-2-specific host pathways that associate with vRNAs.
The team identified 309 host proteins that interact with SARS-CoV-2 RNA during the course of infection.
By comparing the data with ChIRP-MS data for three other positive-sense RNA viruses (Zika, Dengue, and rhinovirus), as well with genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screens, the researchers identified both shared and SARS-CoV-2-specific patterns of RNA–host protein interactions.
For instance, the vRNAs of SARS-CoV-2, Dengue, and Zika are all associated with the Rab proteins RAB10 and RAB2A, which are involved in subcellular trafficking, and these proteins are needed for viral replication and virus-induced cell death.
By contrast, although both human coronaviruses and the flaviviruses require these Rab glycoproteins to produce new infectious virions, the interaction of SARS-CoV-2 vRNA with the translational apparatus and the Sec/Translocon/OST complexes was limited, compared with Dengue and Zika.
“These data suggest that while both form membrane-enclosed replication complexes, flaviviruses may physically leverage the translocon complex, while SARS-CoV-2 leverages other domains of the ERGIC [ER-Golgi intermediate compartment]),” writes the team.
An unexpected finding was that vRNA-binding proteins protected the host
One unexpected finding was that the vast majority of (116/138) of vRNA-binding proteins protected the host from virus-induced cell death, rather than functioning as pro-viral factors.
Most of these antiviral factors were bound to multiple viral families, but the researchers also identified 31 that were specific to SARS-CoV-2.
“These results demonstrate that host cells deploy a broad and diverse array of proteins to physically recognize and counteract viral infection,” say Flynn and colleagues.
SARS-CoV-2 RNA entered the mitochondria
Finally, the researchers also identified a physical connection between SARS-CoV-2 vRNA and host mitochondria, which was validated by electron microscopy data demonstrating changes in mitochondrial shape and size following infection.
The team points out that other viruses, including HIV, have also been reported to enter the mitochondria, suggesting that vRNA can gain access to the mitochondria during infection.
Mitochondria, which are key to maintaining cellular health, play an important role in sensing and signaling during cellular stress, and are vital for innate immune signaling.
“We propose that RNA viruses may follow a distinct logic when causing mitochondrial stress; that is, many viruses may interact with and perturb this organelle, but the precise manner in which stress is caused, and thus signaling occurs, is virus-specific,” write the researchers.
The findings may help to inform future studies
Flynn and colleagues say the study provides an RNA-centric view of the host proteins and RNAs interacting with SARS-CoV-2 RNA during active infection and has identified both shared and SARS-CoV-2-specific patterns of RNA-host protein interactions.
“Altogether, these data provide a comprehensive catalogue of SARS-CoV-2 RNA-host protein interactions, which may inform future studies to understand the mechanisms of viral pathogenesis, as well as nominate host pathways that could be targeted for therapeutic benefit,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Flynn R, et al. Systematic discovery and functional interrogation of SARS-CoV-2 viral RNA-host protein interactions during infection. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.10.06.327445
- Peer reviewed and published scientific report.
Flynn, Ryan A., Julia A. Belk, Yanyan Qi, Yuki Yasumoto, Cameron O. Schmitz, Maxwell R. Mumbach, Aditi Limaye, et al. 2020. “Systematic Discovery and Functional Interrogation of SARS-CoV-2 Viral RNA-Host Protein Interactions during Infection,” October. https://doi.org/10.1101/2020.10.06.327445. https://www.sciencedirect.com/science/article/pii/S009286742100297X.