SARS-CoV-2 is the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic that continues to have a devastating effect on global public health and the economy.
Irene Ong and colleagues say the study findings suggest that the viral membrane (M) protein, particularly the epitope 1-M-24, as well as other highly reactive epitopes should be studied further as potential targets in the development of diagnostics, vaccines, and therapies for SARS-CoV-2.
These epitopes could also be useful for developing diagnostics, vaccines, and therapies for other dangerous coronaviruses that may emerge in the future, they add.
A pre-print version of the paper is available in the server bioRxiv*, while the article undergoes peer review.
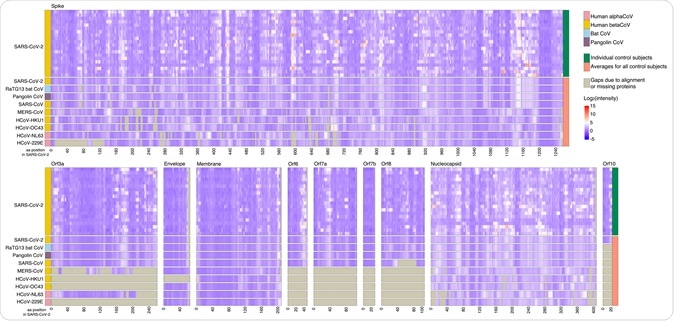
Control sera show reactivity frequently to CCCoVs and rarely to SARS-CoV, MERS-CoV, and SARS-CoV-2. Sera from 20 control subjects collected before 2019 were assayed for IgG binding to the full proteomes of 9 CoVs on a peptide microarray. Viral proteins are shown aligned to the SARS-CoV-2 proteome with each virus having an individual panel; SARS-CoV-2 amino acid (aa) position is represented on the x-axis. Binding was measured as reactivity that was >3 standard deviations above the mean for the log2-quantile normalized array data.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
“Antibody-based options may need expanding”
All coronaviruses have four main structural proteins (spike [S], envelope [E], membrane [M] and nucleocapsid [N]), as well as numerous non-structural proteins and accessory proteins.
In the case of SARS-CoV-2, antibodies against S and N have so far received the most attention.
However, not all infected individuals generate detectable levels of antibodies against these proteins, suggesting that antibody-based options may need to be expanded.
Much less is understood about the antibody responses to other SARS-CoV-2 proteins, although data from studies of other coronaviruses suggest these proteins may be important. For instance, some experimental vaccines for SARS-CoV-1 and the Middle East respiratory syndrome (MERS-CoV) trigger the generation of antibodies against the accessory protein orf8.
Furthermore, previous studies have shown that humoral cross-reactivity occurs between coronaviruses, which could be protective. However, full-proteome cross-reactivity has not yet been investigated.
What did the current study involve?
Now, Ong and colleagues have used a peptide microarray they designed to assess the proteome of SARS-CoV-2 and other human and animal coronaviruses to determine antibody specificity and cross-reactivity between the viruses.
The microarray was used to profile immunoglobulin G (IgG) antibodies among 40 patients who had recovered from COVID-19 and 20 SARS-CoV-2-naive controls.
“We aimed to map the full extent of binding of antibodies induced by SARS-CoV-2 infection and to rank the identified epitopes in terms of likelihood of importance and immunodominance,” writes the team.
The researchers identified 79 B cell epitopes across the structural proteins S, M, N, the polyprotein orf1ab, and the accessory proteins orf3a, orf6, and orf8.
The highest ranking epitope was 1-M-24, which is found in the N-terminus of M.
Patient sera demonstrated high-magnitude reactivity among other epitopes in S, M, N, and orf3a and lower-magnitude reactivity among epitopes in other proteins.
Epitopes with the highest reactivity in S were found in the fusion peptide, while less reactive epitopes were found in the receptor-binding domain.
Four of the epitopes exhibit potent neutralizing activity
Four of the epitopes detected have previously demonstrated potent neutralizing activity, and all of these ranked within the top 10 epitopes in this study.
The team also found that anti-SARS-CoV-2 antibodies cross-reacted with homologous epitopes across the proteomes of other human and animal coronaviruses.
The researchers point out that “hundreds of CoVs [coronaviruses] have been discovered in bats and other species, making future spillovers inevitable.”
The highest number (70) of cross-reactive epitopes were found in the RaTG13 bat betacoronavirus, which is the virus most closely related to SARS-CoV-2 (96% nucleotide identity). This was followed by the pangolin coronavirus (51 epitopes) and SARS-CoV-1 (40 epitopes).
One region corresponding to the SARS-CoV-2 epitope 807-S-26 was cross-reactive across all coronaviruses, and one region corresponding to the SARS-CoV-2 epitope 1140-S-25, was cross-reactive across all betacoronaviruses.
What are the implications of the study?
The researchers say the results suggest that the structural protein M – particularly the 1-M-24 epitope – and the other new epitopes identified here should be explored as important targets in developing improved diagnostics, vaccines, and therapies for SARS-CoV-2 and other dangerous coronaviruses that may emerge in the future.
Furthermore, “the broad cross-reactivity we observed in some homologous peptide sequences may help guide the development of pan-CoV vaccines, especially given that antibodies binding to 807-S-26 and 1140-S-25, epitope motifs cross-reactive across all CoVs and all β-CoVs, respectively, are known to be potently neutralizing,” says the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ong I, et al. The landscape of antibody binding to SARS-CoV-2. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.10.10.334292
- Peer reviewed and published scientific report.
Heffron, Anna S., Sean J. McIlwain, Maya F. Amjadi, David A. Baker, Saniya Khullar, Tammy Armbrust, Peter J. Halfmann, et al. 2021. “The Landscape of Antibody Binding in SARS-CoV-2 Infection.” Edited by Galit Alter. PLOS Biology 19 (6): e3001265. https://doi.org/10.1371/journal.pbio.3001265. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001265.