Syncytia are the large multinucleate cells that have been observed in post-mortem samples of patients who have died from the disease.
Following infection, intracellular trafficking of spike results in its accumulation at the cell surface, where it can induce cell-to-cell adhesion and lead to the formation of multinucleate syncytia.
“Like other viruses, syncytium formation may be advantageous to SARS-CoV-2 by allowing efficient spread between cells that is both rapid and can also evade immune surveillance,” write Sean Munro and colleagues.
The team suggests that syncytia formation by SARS-CoV-2 should be considered as a potential target for therapeutic strategies.
A pre-print version of the paper is available in the server bioRxiv*, while the article undergoes peer review.
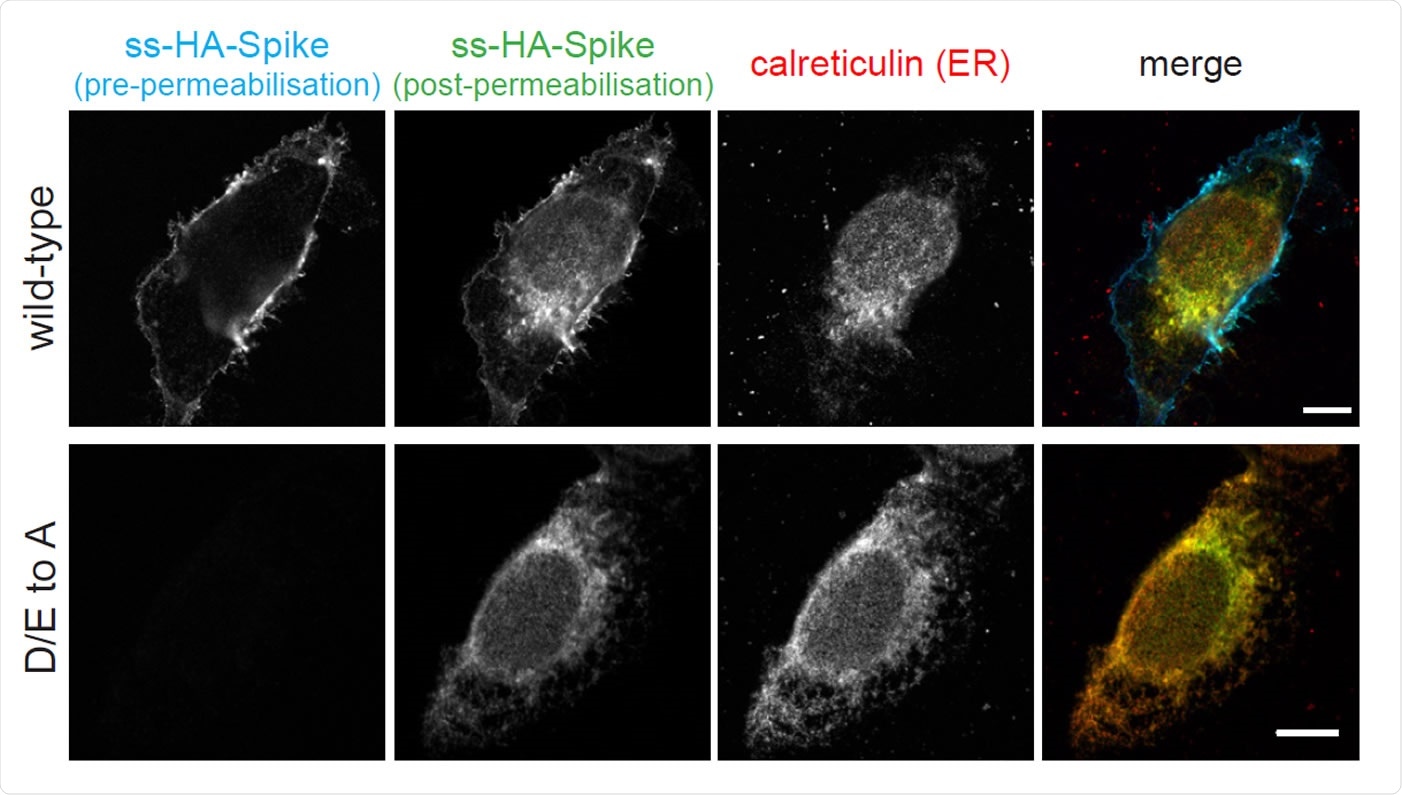
The cytoplasmic tail of the SARS-CoV-2 S protein harbours di-acidic ER export motifs and a suboptimal ER retrieval motif. Micrographs of U2OS cells transiently expressing N-terminally HA-tagged wild-type S and the D/E to A mutant. Cell surface S was initially stained using an anti-HA Alexa Fluor AF647 under non-permeabilising conditions. Cells were subsequently permeabilised and stained with anti-HA AF488 to show the internal S. Scale bars 10 μm. Experiment repeated twice.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The intracellular trafficking of spike
All coronaviruses use a surface spike protein to bind to the host cell receptor angiotensin-converting enzyme 2 (ACE2) and gain viral entry.
Following infection, the spike is inserted into the endoplasmic reticulum (ER) and then trafficked to the ER-Golgi intermediate compartment (ERGIC) and the Golgi, where it undergoes glycosylation and cleavage by Golgi enzymes.
Virions then form through budding into ERGIC and Golgi membranes, and membrane-bound carriers transport them to the cell surface.
“Although SARS-CoV-2 and most other coronaviruses do not bud from the cell surface, some S [Spike] protein is found on the surface of infected cells,” said Munro and team.
The composition of spike
The Spike protein is made up of a large extracellular domain, a transmembrane domain, and a short cytoplasmic tail.
Interactions typically mediate the trafficking of spike through the secretory pathway between its cytoplasmic tail and the coat proteins that form the carriers that transport proteins and lipids between cellular compartments.
To elucidate the intracellular trafficking of the spike in host cells, the researchers applied proteomics to identify the cellular factors that recognize and interact with its cytoplasmic tail.
The team identified several vesicle coat proteins that included subunits of the coatomer complex that form coatomer protein I (COP-I)-coated vesicles and subunits of the COP-II coat that forms ER-to-Golgi vesicles.
In addition to these known interacting factors, the sorting nexin SNX27, which is involved in the recycling of proteins from endosomes to the cell surface, was also identified.
To investigate the specific roles of the different coat proteins, the team mapped the regions they bind to on the Spike protein tail, which is made up of two distinct sections. The membrane-proximal half contains eight cysteines, which, once modified, are embedded in the surface of the lipid bilayer. The distal half of the tail, on the other hand, does not contain cysteines and projects into the cytoplasm.
The interactors mainly bound to the distal region
All of the interactors bound to the distal region of spike, with the exception of SNX27, which only bound to the cysteine-rich membrane region.
On examining the contribution of the COP-II binding site to the subcellular distribution of spike, the team found that acidic residues in this region significantly reduced cell surface expression. Instead, spike accumulated in the ER, suggesting that these residues instruct the movement of a newly-made spike into the secretory pathway.
The incorporation of a COP-I binding site mutation into the full-length spike only caused a small increase in cell surface expression of the protein. On the other hand, the incorporation of two mutations that increased COP-I binding each caused spike to accumulate intracellularly, particularly in the ER.
“Thus, the COP-I binding site in S [Spike] has conserved features that reduce its in vivo efficacy, and so allow it to reach the cell surface,” writes the team.
SARS-CoV-2 Spike does not get endocytosed at the cell surface
Some coronaviruses other than SARS-CoV-2 have been shown to be endocytosed if they reach the host cell surface. In the case of these viruses, endocytosis requires a tyrosine containing a certain motif that resembles the classic Yxxφ signal sequence.
However, the SARS-CoV-2 Spike protein lacks such a motif and, consistent with this, the researchers found that spike simply accumulated at the cell surface and was not endocytosed.
Why might spike accumulate at the cell surface?
Spike that has reached the plasma membrane is then in a position to cause the cell to fuse to adjacent cells and therefore facilitate spread.
“It is known that when at the surface, S can direct cell: cell fusion leading to the formation of multinucleate syncytia,” writes the team.
The researchers say this analysis of the trafficking signals in the Spike protein’s cytoplasmic tail indicates that syncytia formation is not merely an inadvertent by-product of infection, but rather an important aspect of the SARS-CoV-2 replication cycle and a potential cause of pathological symptoms.
“Like other viruses, syncytium formation may be advantageous to SARS-CoV-2 by allowing rapid and efficient spread between cells that is both rapid and can also evade immune surveillance,” write Munro and colleagues.
“Our findings thus argue that syncytia formation by SARS-CoV-2 should be viewed as a potential target for therapeutic strategies,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources