A team of scientists from the United States has found that oral salivary samples are less sensitive than nasal-oropharyngeal swabs for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. However, analysis of oral salivary samples using reverse-transcriptase polymerase chain reaction (RT-PCR) is a reliable approach to identify coronavirus disease 2019 (COVID-19) patients who are more likely to transmit the disease to others. The study is currently available on the medRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Rapid and accurate detection of SARS-CoV-2 infection, along with the strict implementation of control measures, is the primary requirement to contain the COVID-19 pandemic effectively. Since the emergence of the COVID-19 pandemic, nasopharyngeal swabs collected by healthcare workers are considered to be the most effective sampling method to diagnose COVID-19. However, oral salivary samples serve as a potential alternative for diagnostic purposes because of relatively easy sample-collection steps that can be followed by patients themselves. In a hospital setting, patient-collected salivary samples have been shown to have more viral RNA copies than healthcare worker-collected nasopharyngeal swabs.
To establish salivary samples as an alternative diagnostic sample, more studies are required to investigate its sensitivity for diagnosing SARS-CoV-2 infection.
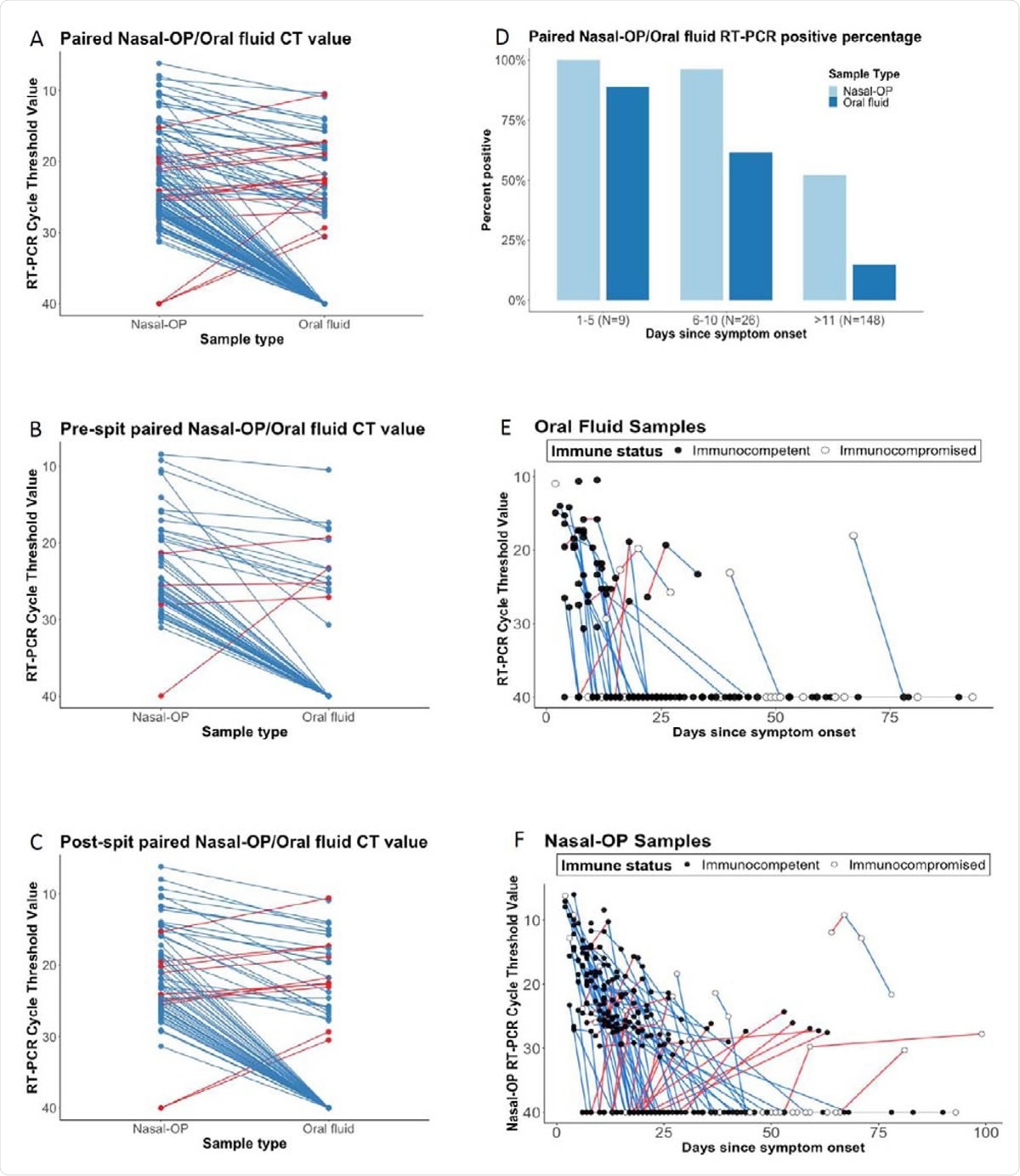
Cycle thresholds (Ct) are plotted for matched nasal-oropharyngeal (OP) swab and oral fluid RT-PCR. Viral burdens that were higher in nasal-OP or oral fluid are shown in blue and red lines, respectively in A) all matched specimens, B) oral fluid only, and C) oral fluid plus the addition of spit. D) The proportion of nasal-OP and oral fluid matched specimens that were positive in participants who are 1-5 days, 6-10, and more than 11 days after symptom onset in samples from participants where the date of symptom onset could be determined. E) Ct values are shown for individual participants over time. Blue lines denote decreasing viral burden whereas red lines represent increasing viral burden with increasing number of days after symptom onset in oral fluid, and F) nasal-OP specimens. RT-PCR Ct values for immunocompetent participants are shown in black filled circles, and for immunocompromised participants in open circles. For figures 1A-C, matched samples that were negative in both sample types were omitted.
Current study design
The current study was designed to investigate the utility of patient-collected oral salivary samples in detecting SARS-CoV-2 infection. The scientists compared the RT-PCR results of 202 matched nasopharyngeal swabs and oral salivary fluid samples (oral crevicular fluid) collected by 67 non-hospitalized COVID-19 patients.
Important observations
The scientists observed that nasopharyngeal swabs had significantly higher viral RNA copies than oral salivary fluids. To improve the sensitivity of salivary samples, the scientists asked the participants to supplement the salivary samples with spit from the back of the mouth and throat. However, no statistically significant improvement in viral RNA detection sensitivity was observed in spit-supplemented salivary samples.
Interestingly, the scientists observed that about 7.7% of pre-spit samples had higher viral RNA copies than nasopharyngeal samples; however, the percentage increased to 14.5% while analyzing post-spit samples. Another important finding was that salivary samples collected at the initial phase of infection (within 5 days) were more likely to provide positive results.
By culturing RT-PCR positive matched samples on VeroE6 TMPRSS2 cells (viral culture to detect infectious virus), the scientists observed that all viral culture-positive samples were also positive in oral fluid RT-PCR. This indicates that oral fluid samples are more sensitive in detecting infectious virus, and thus, can be used to identify COVID-19 patients who are more likely to transmit the disease to others.
Study significance
The current study findings indicate that salivary samples are less sensitive than nasopharyngeal swabs in diagnosing COVID-19. The scientists believe that the sensitivity of salivary samples may depend on the process and timing of collection. Expectorated samples (ejected from the throat or lungs by coughing or spitting) collected in the morning may have a higher viral RNA load because of the enrichment of deeper samples. This indicates that all types of salivary samples may not have comparable detection sensitivity for SARS-CoV-2.
As suggested by the current study, the stage of infection may also influence the sensitivity of salivary samples. Salivary tests may have a higher sensitivity in the first week of infection.
Most importantly, the current study reveals that salivary samples are highly potent in detecting RNA copies of infectious SARS-CoV-2. Despite relatively lower detection sensitivity, the scientists believe that salivary sample testing should be acknowledged widely as a valuable method for COVID-19 diagnosis because of its ability to identify contagious COVID-19 patients.
Moreover, because of the simplicity of the sample collection procedure, patients can easily collect salivary samples without seeking assistance from healthcare workers. This can negate the need for direct patient-healthcare worker interaction.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources