Tracking the trajectory of an outbreak during a pandemic is essential for making key decisions related to public health response measures. Understanding the trends of infection can help decision-makers plan the deployment of public health resources, the need for non-pharmaceutical interventions, and the optimal use of hospital beds for patients and personal protective equipment for health workers.
This is also true for the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. The key epidemiological parameters such as R(t) - the time-varying effective reproductive number – are usually estimated using positive test percentage, daily case counts, or death counts, confirmed by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) testing.
However, the varying symptoms and non-specific incubation periods of coronavirus disease 2019 (COVID-19), the limited availability of testing, and the delay in case reporting or confirmed deaths make these approaches limited and non-reliable in mapping the infection trajectory.
Also, in many countries, changes in case counts are dependent on their changing testing protocols, and hence may not necessarily reflect a real increase or decline in cases. This has significant health, economic, and political ramifications.
Using viral load distribution to interpret epidemic dynamics
Researchers from the Harvard T. H. Chan School of Public Health, Harvard Pilgrim Health Care Institute, and Brigham and Women’s Hospital, Boston, MA; and Vassar College, Poughkeepsie, NY, recently showed that the varying population distribution of Cycle threshold (Ct) values from positive SARS-CoV-2 samples could help infer epidemic dynamics. Their study has been published on the preprint server medRxiv*.
The team showed that the viral load distribution in positive samples at one point in time could help estimate the trajectory of an epidemic accurately. This subverts the need for repeated case count estimations that fluctuate with changes in testing capacity.
“Our results demonstrate that this method can be used to estimate epidemic growth rates based on data collected at a single time point, and independent of assumptions about the intensity of testing.”
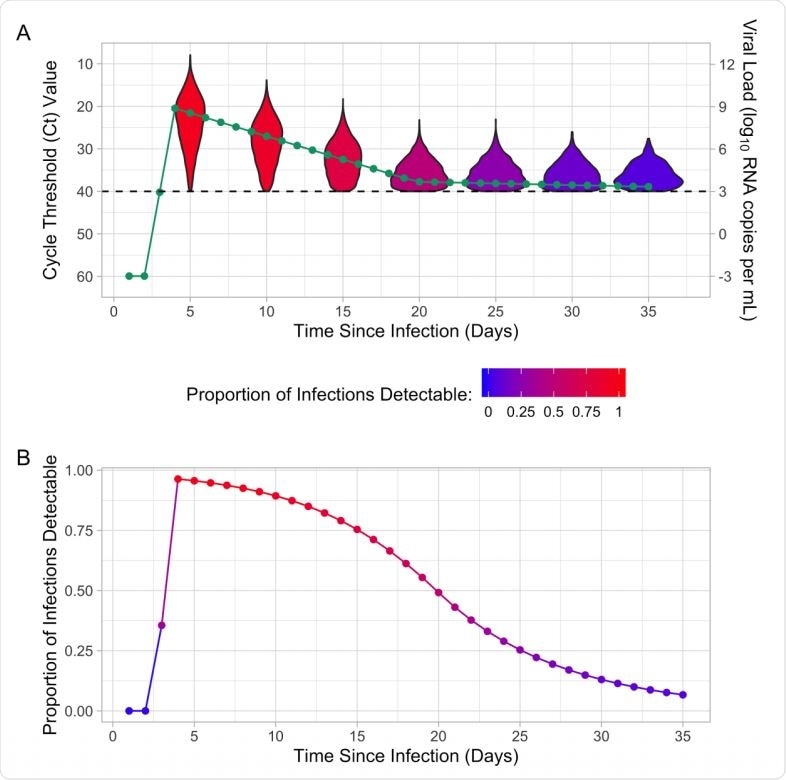
(A) Mean cycle threshold (Ct) value, mean viral load, and distribution of Ct values for detectable infections by time since infection. (B) Proportion of infections that are detectable by time since infection for the population-level Ct distribution. The proportion of infections that are detectable is indicated by the color of the violin plot and the proportion detectable line. The dashed line indicates the limit of detection (Ct value of 40 or viral load of 3).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Novel approach helps estimate the position of a community in the epidemic curve
Virologic testing for the SARS-CoV-2 virus has been central to the COVID-19 pandemic response, but understanding the trends in incidence and positive tests and hence the epidemic trajectory is limited by variations in testing practices.
The researchers identified a link between the population-level, cross-sectional distribution of Ct values, and the epidemic growth rate in the study. This demonstrated that the change in skewness and median of detectable Ct values are purely a mathematical epidemiologic rule without any change in individual viral load kinetics or testing.
Although the individual-level variation in measurement can complicate Ct values' interpretation in the clinical setting, the authors hypothesize that population-level findings reflect the underlying epidemic dynamics.
In agreement with these theoretical findings, they also observed a robust relationship between R(t) and Ct distribution in positive specimens analyzed in Massachusetts. Their novel method can be used to estimate the position of a community in the epidemic curve, as defined by the growth rate, based on a single cross-sectional analysis of virologic testing data.
They used the observed links to develop a novel method that helps accurately infer the epidemic growth rate using the Ct distribution values observed at a single cross-section in time. This, unlike estimates based on case counts, is less prone to biases caused by test result delays and changing testing protocols.
According to the authors, their findings show that by incorporating viral loads into public health data streams, a new approach can be devised that allows for real-time resource allocation and assessment of outbreak mitigation strategies, even in the absence of repeat incidence data. They think that it would help if testing centers regularly report ct values or similar viral load data to public health officials, and these data are incorporated into outbreak monitoring systems.
The researchers hope that future research studies will evaluate how to incorporate these data into an overall inferential framework for real-time epidemic trajectory monitoring.
“By using Ct values to determine the growth rate of incident cases and using virologic positivity rates to assess prevalence of infection, as well as potentially incorporating measured covariates and symptom status, a richer picture of the path and likely future of an outbreak can be realized from one or more virologic surveys.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Estimating epidemiologic dynamics from single cross-sectional viral load distributions James A Hay, Lee Kennedy-Shaffer, Sanjat Kanjilal, Marc Lipsitch, Michael J Mina medRxiv 2020.10.08.20204222; doi: https://doi.org/10.1101/2020.10.08.20204222, https://www.medrxiv.org/content/10.1101/2020.10.08.20204222v1
- Peer reviewed and published scientific report.
Hay, James A., Lee Kennedy-Shaffer, Sanjat Kanjilal, Niall J. Lennon, Stacey B. Gabriel, Marc Lipsitch, and Michael J. Mina. 2021. “Estimating Epidemiologic Dynamics from Cross-Sectional Viral Load Distributions.” Science, June. https://doi.org/10.1126/science.abh0635. https://www.science.org/doi/10.1126/science.abh0635.