Severe acute respiratory coronavirus 2 (SARS-CoV-2) belongs to the beta-coronavirus family. Coronaviruses are positive-sense, single-stranded RNA viruses, and have one of the largest genomes among RNA viruses.
Out of the 7 known coronaviruses that have infected humans so far, 4 (HCov-229E, HCoV-OC43, HCoV-NL63, CoV-HKU1) can cause seasonal, mild respiratory symptoms in healthy people. In contrast, the SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) are closely related to SARS-CoV-2 and have caused the 2002-2003 SARS and 2012 MERS epidemics, respectively. The current coronavirus disease of 2019 (COVID-19) pandemic is a global public health emergency with over 38 million cases and 1.08 million deaths worldwide to date.
The SARS-CoV-2 spike (S) glycoprotein
The surface of the SARS-CoV-2 virus has a spike (S) glycoprotein, which is a large homo-trimeric glycoprotein that forms a crown at the virus capsid surface. Each S protomer is further divided into two domains - S1 and S2 – that are demarcated by a furin cleavage site. The S1 domain is responsible for recognition and binding to the angiotensin-converting enzyme 2 (ACE2) receptor in the host cell. At the same time, the S2 domain directs the fusion of viral and host cell membranes and also undergoes significant conformational changes upon furin cleavage.
Previous studies had reported the central role of the receptor-binding domain (RBD) domain dynamics between “closed” and “open” conformations for the recognition and binding to the ACE2 receptor of the host cell. Thus, the S protein of SARS-CoV-2 is the target of most vaccine development efforts to bring the COVID-19 pandemic under control.
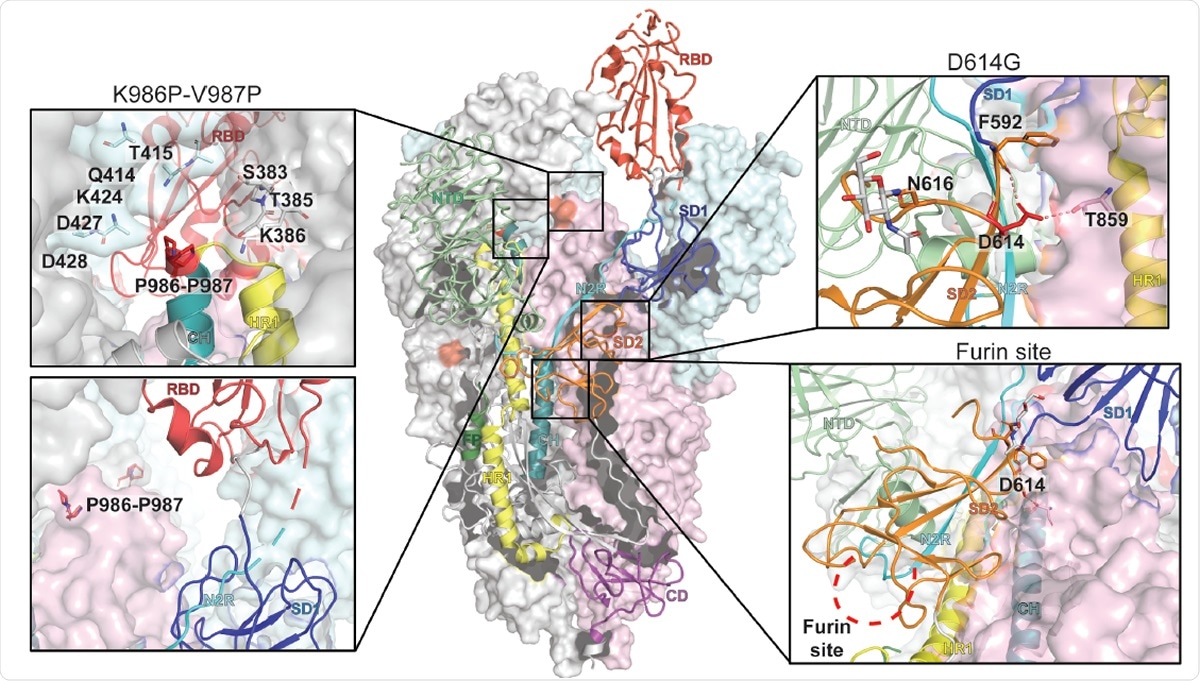
Representation of the trimeric SARS-CoV-2 spike ectodomain with one RBD-up in a prefusion conformation (PDB ID 6VSB). The S1 domain on an RBD-down protomer is shown as pale green molecular surface while the S2 domain is shown in pale red. The subdomains on an RBD-up protomer are colored according to panel A on a ribbon diagram. Each inset correspond to the spike regions understudy and are highlighted in red on the trimeric structure (K986P-V987P, D614G and the furin protease cleavage site).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
D614G mutation of the S glycoprotein
Despite a low mutation rate of the SARS-CoV-2, mutated viruses with D614G substitution in the S protein were isolated early in the pandemic and are now the most dominant form of SARS-CoV-2 worldwide. The D614G mutation of the S glycoprotein has been linked to increased infectivity of the virus in many studies. Cryo-electron microscopy (cryo-EM) structures of the S protein ectodomain shows that D614 is a surface residue around the furin cleavage site.
“The D614G mutation of the S protein has been associated in numerous reports with increased fitness or infectivity of the virus.”
Researchers from the Duke Human Vaccine Institute and Duke University, Durham NC, USA, recently analyzed the D614G mutation in SARS-CoV-2 in the context of a soluble S ectodomain construct. Their study has been published in the preprint server bioRxiv*.
They performed Cryo-EM, proteolysis, and antigenicity experiments and found altered conformational dynamics that led to enhanced furin cleavage efficiency of the G614 variant. Moreover, furin cleavage changed the conformational dynamics of the RBD in the G614 S ectodomain. This demonstrates an allosteric effect on the RBD dynamics caused by changes in the SD2 region. This region houses the furin cleavage site as well as residue 614.
Spike conformational dynamics and allostery of SARS-CoV-2 could aid vaccine design
In this study, the researchers identified the SD2 domain in the S protein of SARS-CoV-2 as a conformational anchor spatially interspersed between 2 highly mobile regions, while itself being relatively invariant in conformation. The findings suggest a role for the SD2 domain in preventing premature triggering in the SARS-CoV-2 S protein and in other key downstream events such as ACE2 receptor engagement and TMPRSS2 protease cleavage.
In addition to the D614G mutation, the SD2 subdomain also has the furin cleavage site. Furin cleavage is a significant step for the S protein and is essential for viral infection as well as transmission. According to the team, their study provides evidence that D614G mutation increases susceptibility of the SARS-CoV-2 S ectodomain to furin cleavage, thus enhancing the possibility that it increases fitness and transmissibility of viruses with the D614G mutation.
According to the researchers, they were able to show that the S ectodomains of SARS-CoV-2 with or without the K986P, V987P mutations had similar structures, stability, and antigenicity. Also, cryo-EM structures showed the allosteric effect that changes at the S1/S2 junction have on RBD dynamics. Thus they claim that the results of the study explain the spike conformational dynamics and allostery of the SARS-CoV-2 virus and could have significant implications for the development of a vaccine for COVID-19.
“We provide evidence in this study that the D614G mutation enhances susceptibility of the SARS-CoV-2 S ectodomain to furin cleavage, thus raising the possibility that this is a contributor to increased fitness and transmissibility of D614G isolates.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
D614G mutation alters SARS-CoV-2 spike conformational dynamics and protease cleavage susceptibility at the S1/S2 junction Sophie Gobeil, Katarzyna Janowska, Shana McDowell, Katayoun Mansouri, Robert Parks, Kartik Manne, Victoria Stalls, Megan Kopp, Rory Henderson, Robert J Edwards, Barton F Haynes, Priyamvada Acharya bioRxiv 2020.10.11.335299; doi: https://doi.org/10.1101/2020.10.11.335299, https://www.biorxiv.org/content/10.1101/2020.10.11.335299v1
- Peer reviewed and published scientific report.
Gobeil, Sophie M.-C., Katarzyna Janowska, Shana McDowell, Katayoun Mansouri, Robert Parks, Kartik Manne, Victoria Stalls, et al. 2021. “D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction.” Cell Reports 34 (2): 108630. https://doi.org/10.1016/j.celrep.2020.108630. https://www.cell.com/cell-reports/fulltext/S2211-1247(20)31619-3.