The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus that emerged in Wuhan, China, in late 2019 has caused the ongoing coronavirus disease 2019 (COVID-19) pandemic, which has affected more than 39.8 million lives globally and claimed over 1.11 million deaths to date. While extensive sequencing efforts are ongoing to understand this virus's evolution, several studies with SARS-CoV-2 sequencing data show different allele frequencies of the virus in the same patient, a phenomenon called heteroplasmy.
The most probable explanation for this intra-patient heterogenic viral reads is the existence of multiple viral strains. Recombination is an unlikely explanation because the chances of the virus being functional after disassembly inside the host cell and reassembly into a virion having a different sequence are pretty low. While there is evidence of multiple lineages of SARS-CoV-2 virus in the same COVID-19 patient, no evidence of sublineages recombining in the same patient is available to date.
Clinical implications of heteroplasmy
Multiple strains of a virus infecting the same patient have huge clinical implications in epidemiology, treatment, and controlling the pandemic. Variations in viral strains can indicate different transmissibility levels, different drug resistance mechanisms, varying responses to treatment, and explain the wide variety of symptomology. Given the significance of this in treatment and vaccine development, it is imperative that the more research focuses on heteroplasmy of SARS-CoV-2.
Researchers from the IBM Research, T.J. Watson Research Center, NY, USA, recently presented a common methodological framework to interpret the phylogenomics from genomic data for multiple diseases, including COVID-19 and cancer. Their work is published on the preprint server bioRxiv*.
In the case of cancer, the tumor heterogeneity in a patient indicates intra-patient heteroplasmy, and the absence of recombination in tumor cells is an accepted assumption. The researchers hypothesize that just like the different frequencies of the genomic variants of a tumor indicates multiple tumor clones and offers a handle to infer them computationally, the different variant frequencies in viral genomic reads offer the means to compute the multiple co-infecting sublineages.
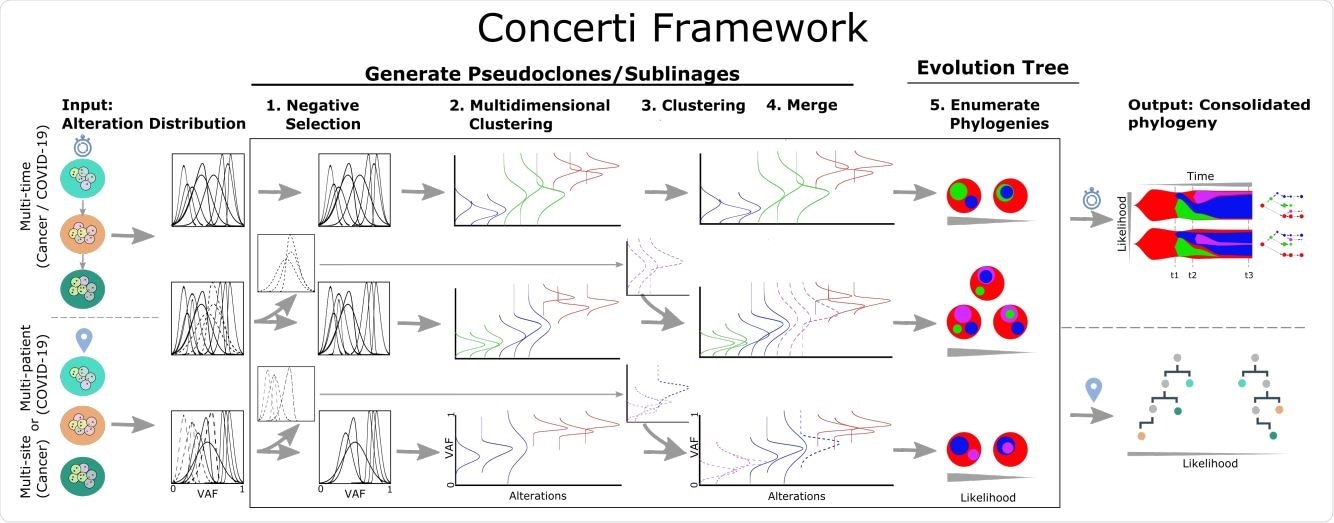
Schematic of the Concerti Framework. Given a set of multi-patient (COVID-19) or multi-site, multi-time (cancer) genomic samples, the algorithm analyzes the underlying alteration frequency distribution as input and performs a (1) negative selection to filter appearing alterations. A (2) multidimensional clustering is done to identify pseudoclones/lineages that will then be enriched by a (3) single sample clustering that (4) merges alterations that were initially negatively selected. (5) All potential phylogenies are generated and assessed for compatibility. Finally the set of consolidated phylogenetic structures over time or site are output with likelihood scores.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
An algorithm for understanding evolutionary phylogenies
The study describes a computational framework called Concerti to infer phylogenies in both the above scenarios. To demonstrate the accuracy of this algorithm, the researchers reproduced some previously known results in both scenarios. They also identified a novel potential parallel mutation in the SARS-CoV-2 virus and uncovered new clones having therapy-resistant mutations in the context of cancer.
According to the researchers, Concerti's ability to extract and integrate information from multiple points, sites, times, or samples makes it possible to discover phylogenetic trees that capture the spatial and temporal heterogeneity. These phylogeny models can directly impact therapeutics as they can highlight the "birth" of clones that may harbor mechanisms of treatment resistance, "death" of subclones with drug targets, and the acquisition of functionally relevant mutations in clones that may seem clinically irrelevant.
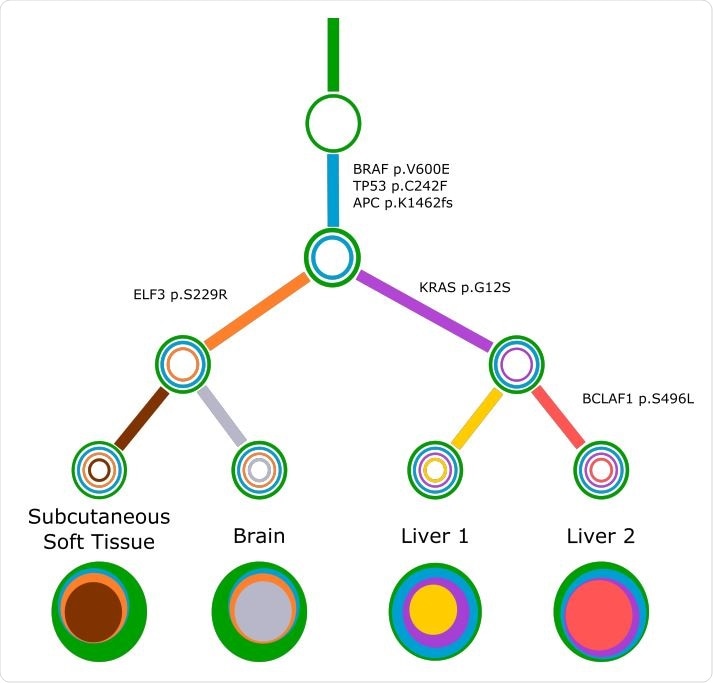
Concerti tumor evolution tree T for patient GI1. Tumor evolution tree T for colon cancer patient GI1 multi-site data. The edges of the T are labeled by the known cancer genes and the colors denote the distinct pseudoclones estimated by Concerti. Leaf nodes represent each of the distinct lesion sites. The single site trees T are shown at the bottom as stacked discs and the sizes are proportional to the prevalence values.
The team demonstrated how Concerti could be applied to any genomic sequencing dataset with different allele frequencies, be it cancer or SARS-CoV-2, and how the results provided by the algorithm can have significant disease-specific clinical implications.
"We demonstrate in this paper how Concerti can be applied to any genomic sequencing dataset with varying allele frequencies, whether it be cancer or the new SARS-CoV-2 virus causing the COVID-19 pandemic, and the results can have profound disease-specific clinical implication."
Specific integration of multi-point data could improve treatment response
Identifying the presence of many viral strains in a single host can profoundly impact treatment approaches, vaccine development efforts, and infection mitigation strategies. Concerti data for COVID-19 patients shows the ability to identify viral strains based on different allele frequencies and thus discover the presence of new homoplasies. The researchers believe that the results provided by Concerti effectively addresses crucial challenges faced by researches in the development of therapeutics and vaccines.
With cancer, accurate monitoring of tumor evolution over the disease course can help identify new drug targets and therapeutic methods that could stabilize this disease and manage the pressures of treatment exposure and tumor environment changes. The study results highlight how specific integration of multi-point data by Concerti could facilitate more optimized and locally targeted treatment plans for better treatment responsivity.
"Concerti's results address the overwhelming challenges researches face when developing 396 therapeutics and may help facilitate the key to effective vaccine development."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources