The novel coronavirus was first identified in patients presenting with pneumonia of unknown cause in Wuhan, China, in late December 2019. The cases were caused by the new pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has now spread worldwide. Over 40 million cases of SARS-CoV-2 infection have been reported, resulting in over 1 million deaths. Mitigation of this infection requires effective therapeutic drugs immediately.
To this end, understanding the virus and its mechanisms is crucial. Recently inactivation of the virus to contain its spread has become the objective of researchers across various disciplines. Treatment methods employing chemical, biological, and physical strategies are being used to slow this viral spread. While SARS-CoV-2 is reported to be highly stable at low temperatures (i.e., 4 °C up to 14 days), it is known to be sensitive to heat.
Didac Martí et al., from the Polytechnic University of Catalonia, in a recent bioRxiv* preprint research paper, explore the effect of the temperature on the molecular structure of the SARS-CoV-2 spike glycoprotein. The study specifically focuses on the spike glycoprotein's molecular structural integrity at different temperatures (within 25 ºC and 100 ºC) using atomistic computer simulations.
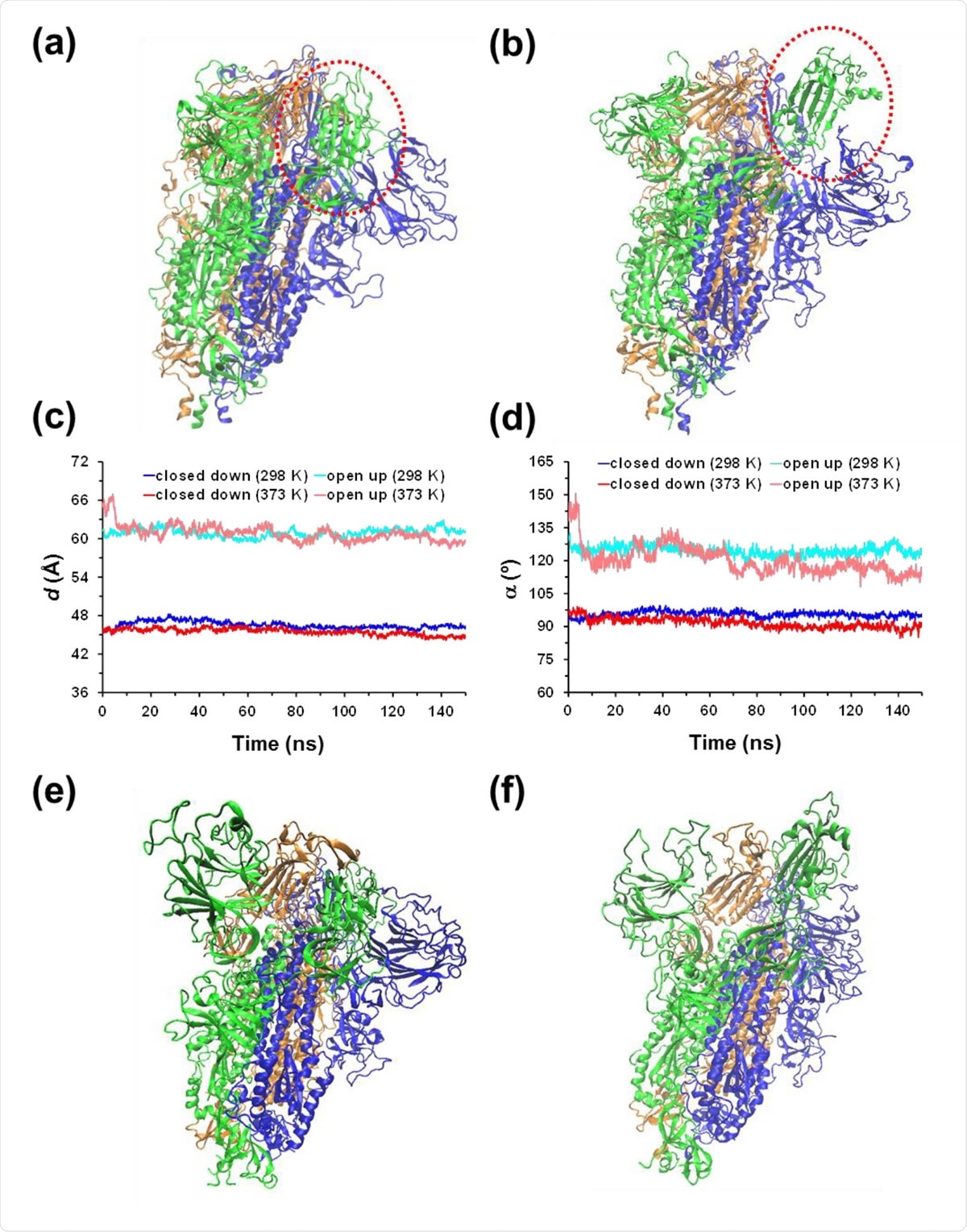
(a) Closed down and (b) open up conformational states of the SARS-CoV-2 homotrimeric spike protein. The main difference between such two states is marked by the red dashed circle. For the MD simulations conducted at 298 and 373 K, the temporal evolution of the geometric parameters used to identify the conformational states of the protein: (c) distance (d) between the center of mass of the RBD from chain B to the center of mass of three monomers; and (d) hinge angle (α) formed by the center of mass of the RBD of chain B, the center of mass of the rest of chain B, and the center of mass of the first residue after the RBD. For the (e) closed down and (f) open up states, structure of the spike protein at the end of the MD simulation at 373 K.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Both the closed down and open up states - which determine the accessibility to the receptor-binding domain (RBD) - were considered. The study results suggest that the drastic change in the hydrogen bonding (H-bond) inactivates the virus, and the intact salt bridges in the protein retain the macrostructure of the spike.
The SARS-CoV-2 virus uses the spike glycoprotein, a homotrimer with three monomers, named chain A, B, and C, to bind to host cellular receptors (human angiotensin-converting enzyme 2 (ACE2). This triggers a cascade of reactions resulting in viral entry into the host.
In this study, the atomistic Molecular Dynamics (MD) simulations were conducted on this homotrimeric protein in an aqueous solution. Both the closed and open conformational states of the protein were studied. The closed down state is when the receptor binding for interacting with ACE2 is hidden; the open up state is when the receptor-binding motifs are exposed.
MD is a computational tool that captures time-dependent conformational changes at various conditions. It calculates the interatomic forces through a solvent, providing the underlying dynamics of the specific molecule.
The Protein Data Bank (PDB) is the source for the atomic coordinates of the homotrimeric spike glycoprotein of SARS-CoV-2 in both conformational states.
Although this study does not elaborate on the different conformations' energetics, the authors observe that the closed down conformational state is stabilized. This stability order is also observed in experiments. The thermally-induced structural distortions are more pronounced for the open state.
The authors report that temperature affects the spike glycoprotein, which renders the virus inactive after a certain threshold point. The temperature changes induce a remodeling of the internal hydrogen-bonding network. Significantly, this affects the recognition functionality of the receptor-binding domain.
Conversely, salt bridge topology remains mostly unaltered. The macrostructure of the spike is thus preserved at high temperatures because of the retained salt bridges. A detailed analysis of the structural conformation's temporal evolution at different temperatures is discussed in this study. The proposed mechanism has important implications for engineering new approaches to inactivate the SARS-CoV-2 virus.
MD is the best methodology to discover atomic interactions in silico to unveil a molecular mechanism for the virus inactivation; performing this study experimentally in a physical lab is a challenge. However, the study's insights contribute enormously to the development of new strategies to disable the virus's functional sites - enabling effective mitigation steps to control the spread of infection.
The authors call for new strategies to inactivate the SARS-CoV-2 using chaotropic agents and surfactants or through physical treatments to selectively target such labile hydrogen bond structures.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources