Researchers have long established that the binding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to the host angiotensin-converting enzyme 2 (ACE2) is essential for the virus to gain entry to and infect the host cell. A new study published in the reputed journal Science in October 2020 reports the potential therapeutic value of the second type of host transmembrane receptor, called neuropilins, which attach to the spike protein of SARS-CoV-2 and may enhance its infectivity.
In this breakthrough study, the research groups in Bristol's Faculty of Life Sciences, Professor Peter Cullen from the School of Biochemistry; Dr. Yohei Yamauchi, Associate Professor, and virologist from the School of Cellular and Molecular Medicine.
Dr. Boris Simonetti, a senior researcher in the Cullen lab, used multiple approaches to discover that SARS-CoV-2 recognizes a protein called neuropilin-1 on the surface of human cells to facilitate viral infection.
Yohei, Boris, and Pete explained: "In looking at the sequence of the SARS-CoV-2 Spike protein we were struck by the presence of a small sequence of amino acids that appeared to mimic a protein sequence found in human proteins which interact with neuropilin-1.
This led us to propose a simple hypothesis: could the Spike protein of SARS-CoV-2 associate with neuropilin-1 to aid viral infection of human cells? Excitingly, in applying a range of structural and biochemical approaches we have been able to establish that the Spike protein of SARS-CoV-2 does indeed bind to neuropilin-1.
"Once we had established that the Spike protein bound to neuropilin-1 we were able to show that the interaction serves to enhance SARS-CoV-2 invasion of human cells grown in cell culture.
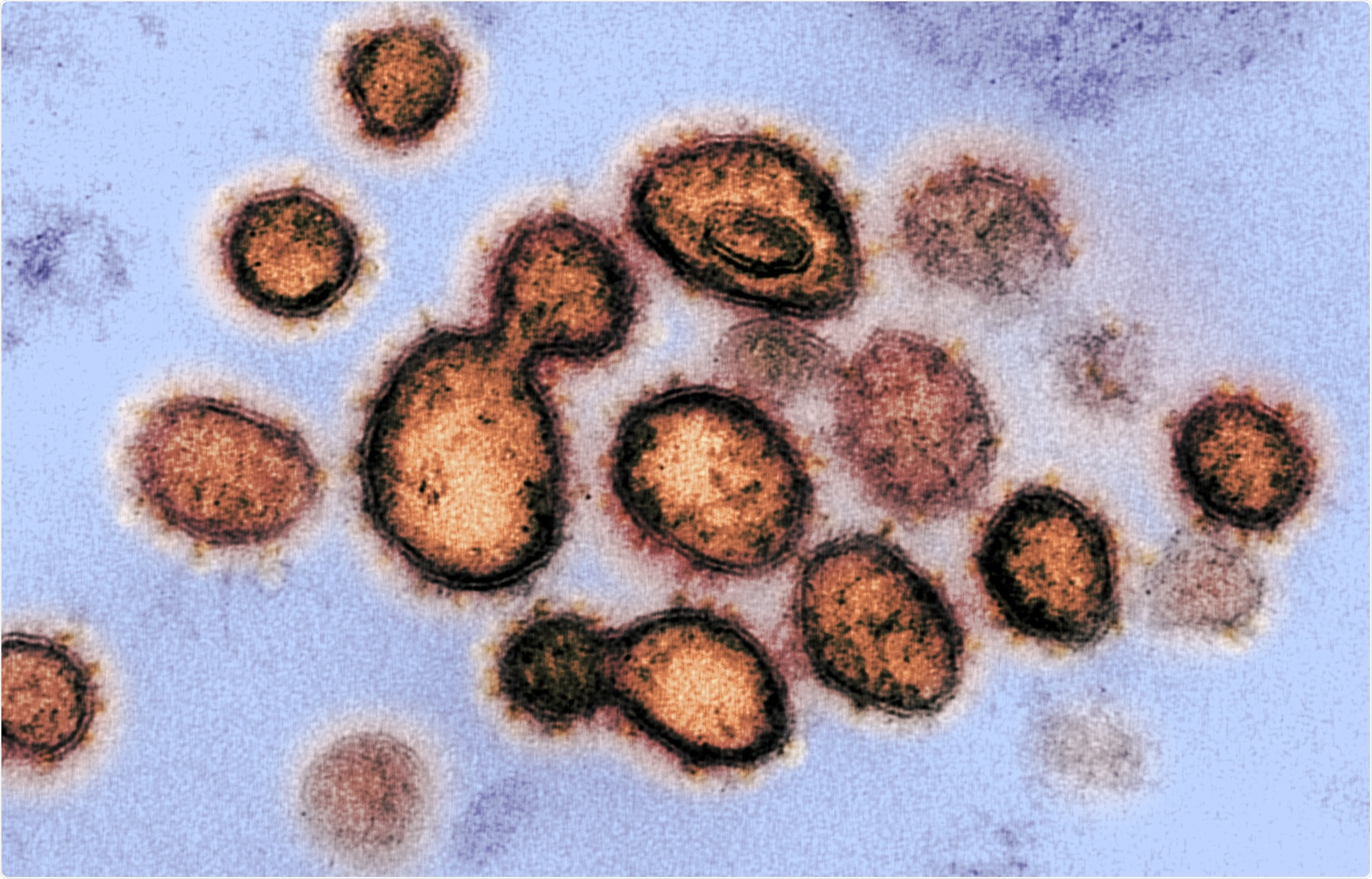
This transmission electron microscope image shows SARS-CoV-2, the virus that causes COVID-19, isolated from a patient in the U.S. Virus particles are shown emerging from the surface of cells cultured in the lab. The spikes on the outer edge of the virus particles give coronaviruses their name, crown-like. Image captured and colorized at NIAID's Rocky Mountain Laboratories (RML) in Hamilton, Montana. Credit: NIAID
The Spike Cleavage Site
The COVID-19 pandemic has caused an immense number of cases, numbering over 42.9 million, with well over a million deaths so far worldwide. The viral agent, SARS-CoV-2, enters the host cell via its spike protein binding to the ACE2 receptor on the cell. The spike protein comprises two subunits, S1 and S2, and has a polybasic furin cleavage site at the interface of the two. This allows a proprotein convertase, or furin, of the host cell to cleave the spike once it has engaged the receptor.
The S1 and S2 subunits are still associated non-covalently, and the S2 appears to be further primed by the serine protease TMPRSS2. Furin cleavage of the spike protein leads to enhanced viral infectivity. Furin inhibition, or deletion of the polybasic RRAR site, reduces viral entry.
The Neuropilin Factor
The S1 subunit has a C-terminal amino acid sequence which follows the C-end rule (CendR). Peptides of this type attach to Neuropilin-1 (NRP1) and NRP2, these being transmembrane receptors that affect many diverse processes of the body. The current study was aimed at exploring the connection between these two.
The researchers used a GFP-labelled S1 construct expressed in ACE2-positive cells in culture. They found that the S1 protein was bound to both ACE2 and NRP-1 in the GFP-S1 cells but not the GFP-S1 ΔRRAR cells, which had a deletion of the furin cleavage site. S1 binding to a similar degree was also observed with NRP2. Some binding was also observed to the deletion mutant showing that the neuropilins did interact with the S1 independently of the CendR residues.
Absence of NRP1 Reduces Infectivity
In cells that lacked NRP1 but expressed ACE2, challenge by the whole virus was found to result in lower levels of infection. Cells lacking both were not infected at all. In another cell assay, treatment with the suppressor shRNA that prevented NRP1 expression again reduced the virus's level of infection. The absence of this protein still allows the virus to bind to the cell surface, but its internalization is cut by half. Thus, NRP1 is shown to be vital for the entry of the virus and its infection of the cell. Their results also supported the existence of a CendR-independent mechanism of enhanced infection mediated by NRP1.
SARS-CoV-2 infection was markedly greater in NRP-positive cells, but in the presence of the T316R mutation, no increase was observed, indicating this change reduced the association of NRP1 with the S1 subunit. They found that monoclonal antibodies targeting the NRP1 protein bound to the CendR-binding pocket expressed on the cell, with high specificity, and reduced the ability of SARS-CoV-2 to infect the cell. Incubation with soluble ACE2 also prevented SARS-CoV-2 infection, indicating the validity of the previous finding.
The use of a small molecule NRP1 inhibitor EG00229 also showed that it could effectively inhibit S1-NRP1 binding by binding to the b1 CendR binding pocket of the NRP1. Thus, this presents a therapeutic target to reduce the infectivity of the virus in human cells.
How Neuropilins Increase Viral Infectivity
Viral entry is preceded by spike priming by host proteases. The CendR sequence produced by the spike protein's cleavage at the S1/S2 interface binds to the neuropilins at the cell surface. This is not essential for the viral attachment to the cell surface, but it does enhance viral entry and cell infection.
The researchers are not sure how neuropilins cause increased levels of viral infection. However, they postulate that this may be due to neuropilins' known ability to mediate macropinocytosis, where the cell engulfs a large pocket of the cell membrane and the attached molecules and particles allowing the entry of CendR-bound molecules into the cell.
Both NRP1 and NRP2 are known to be upregulated in lung tissue samples from COVID-19 patients. Therefore, the binding of NRP1 to the S1-CendR motif may be one reason why SARS-CoV-2 is more infective than SARS-CoV. This may be targeted in future treatments for this disease.