Researchers in the U.S. have looked to expand the knowledge of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoprotein's evolution, function, and interactions with host factors to help researchers develop effective vaccine immunogens and treatments.
SARS-CoV-2 is the causative agent of COVID-19 disease, currently raging across the world. The virus has now infected more than 43.89 million individuals and claimed over 1.16 million lives. The study titled, "Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects," is released on the preprint server bioRxiv* in October 2020.
Background
Coronaviruses are enveloped viruses with a positive-strand RNA as its genetic material. These infect the respiratory and digestive tracts of animals and humans. Two betacoronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV-1) and Middle Eastern respiratory syndrome coronavirus (MERS-CoV), caused deadly but self-limited outbreak in humans in 2002 and 2013, respectively. The current SARS-CoV-2 pandemic leads to severe respiratory disease in humans with a 3 to 4 percent death rate.
What happens when the virus hits a host cell?
SARS-CoV-2 contains a spike (S) glycoprotein trimer and a Class I fusion protein. This is responsible for the virus's entry into the host cell and leads to the pathological effects on the cell or cytopathic effects. It is broken into S1 and S2 glycoproteins. This study attempted to see the contributions of several S glycoprotein features on the effects and the differences in the other related coronaviruses. The SARS-CoV-2 spike glycoprotein (S gp) is one of the targets of a vaccine that could prevent COVID-19.
Among the 2 subunits, S1 contains a receptor-binding domain (RBD) that binds to the angiotensin-converting enzyme 2 (ACE2) receptor. The S2 subunit has a fusion peptide and two heptad repeat regions (HR1 and HR2). These regions allow the fusion of the viral membrane and the cell membrane of the host. Cleavage of S1 and S2 takes place with the help of a protease enzyme produced by the host cell called Cathepsin B/L, TMPRSS2. Then the HR1 and HR2 regions of S2 form a stable six-helix bundle that brings together the viral and cell membranes and allows entry of the virus.
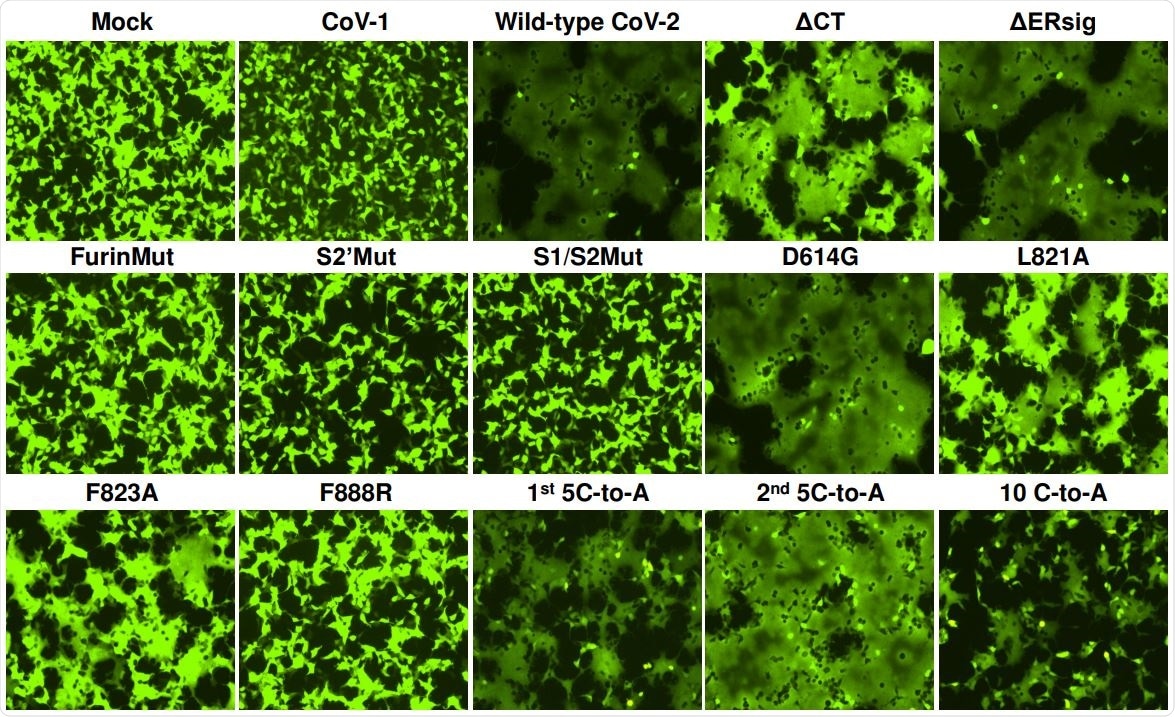
293T-ACE2 cells were transfected either with a plasmid expressing enhanced green fluorescent protein (eGFP) alone (Mock) or with the eGFP-expressing plasmid and a plasmid expressing SARS-CoV-1 S gp or wild-type or mutant SARS-CoV-2 S glycoproteins. After 24 hours, the cells were examined under a fluorescent microscope. The results shown are representative of those obtained in two independent experiments.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Differences in S glycoprotein between SARS CoV1, MERS CoV, and SARS CoV2
- A diversity in S gp alters the interaction between the host cell and the virus and affects coronaviruses' pathogenicity. The authors explained that the S gp of SARS-CoV-2 is 79.6 percent similar to that of SARS-CoV-1 but has some differences. It has furin bases between its S1 and S2 junction. This cleavage site is also seen in MERS-CoV S gp.
- Compared with the original SARS-CoV-2 found in Wuhan, China, the present SARS-CoV-2 variants have a glycine residue instead of aspartic acid at the 614 (D614G) position on the S1 and a C-terminal domain (CTD2). The authors write that this D614G change is associated with a faster and higher replication rate of the virus.
- The SARS viruses have a 97 percent similar cysteine-rich S2 endodomains. SARS-CoV-2, in addition, has cysteine residue in its cytoplasmic tail. The S2 endodomain of SARS-CoV-1 can potentially influence the S gp function and localization and assembly of the virus in the endoplasmic reticulum-Golgi intermediate complex (ERGIC).
Methods
The study by researchers from Dana-Farber Cancer Institute, Harvard Medical School, and the University of Alabama at Birmingham was conducted to evaluate the phenotypes of SARS-CoV-2 S gp mutants and check on the two proposed cleavage sites at the S1/S2 junction and the polymorphic residue Asp/Gly 614. They also studied the fusion peptide in the S2 ectodomain and the ERGIC retention signal, and the palmitoylated cysteine residues in the S2 endodomain.
For this study, they looked at the S gp expression on the genetic makeup of the virus and the "association, glycosylation, and incorporation" of these sequences onto lentivirus and vesicular stomatitis virus (VSV) and other SARS-CoV-2 virus-like particles (VLPs).
When incorporated into other viruses, they noted if the S glycoprotein variants were capable of making them infective and the levels of stability and sensitivity to inhibition of viruses. They also checked if the S gp variants could promote cell-cell fusion to form deadly combinations or syncitia. TMPRSS2 and matrix metalloproteases were checked to see if they could contribute to the efficiency of this process. Matrix metalloprotease inhibitors were checked to see if they could block S-mediated cell-cell fusion in TMPRSS2.
What was found?
The researchers noted that this furin cleavage site of the SARS-CoV-2 S glycoprotein reduced the virus stability and infectivity but raised its ability to form lethal syncytia. They noted that the D614G change seen in the present strains of SARS-CoV-2 restores its infectivity and enhances its affinity towards the ACE2 receptor and susceptibility to an antibody-containing serum. The D614G change also strengthens the association of the S1 subunit with the trimer.
The team writes, "Apparently, two unique features of the SARS-CoV-2 S glycoprotein, the furin cleavage site, and D614G, have evolved to balance virus infectivity, stability, cytopathicity, and antibody vulnerability."
They also noted that the endodomain (cytoplasmic tail) of the S2 subunit decreased the virus entry or syncytium formation. They also found that the protease inhibitors prevent proteases from breaking the S gp and suppress the S glycoprotein function. Matrix metalloprotease inhibitors also decrease the S-mediated cell-cell fusion but do not affect the cell's virus entry. Authors wrote, "Synergy between inhibitors of matrix metalloproteases and TMPRSS2 suggest that both proteases can activate the S glycoprotein during the process of syncytium formation".
Importance and implications
SARS-CoV-2 S glycoprotein-host cell interactions could help determine the virus's risk of transmission and pathogenicity, write the researchers. The authors explain that since the S gp is the primary vaccine target and also a target for the neutralizing antibodies formed during an active infection, this study provides an important insight into this protein. The authors concluded, "Knowledge of the spike glycoprotein evolution, function, and interactions with host factors will help researchers to develop effective vaccine immunogens and treatments." They identified two metalloproteases that could block S-mediated cell-cell fusion, which could help prevent infection and its mitigation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects Hanh T. Nguyen, Shijian Zhang, Qian Wang, Saumya Anang, Jia Wang, Haitao Ding, John C. Kappes, Joseph Sodroski bioRxiv 2020.10.22.351569; doi: https://doi.org/10.1101/2020.10.22.351569, https://www.biorxiv.org/content/10.1101/2020.10.22.351569v1.full
- Peer reviewed and published scientific report.
Nguyen, Hanh T., Shijian Zhang, Qian Wang, Saumya Anang, Jia Wang, Haitao Ding, John C. Kappes, and Joseph Sodroski. 2021. “Spike Glycoprotein and Host Cell Determinants of SARS-CoV-2 Entry and Cytopathic Effects.” Edited by Rebecca Ellis Dutch. Journal of Virology 95 (5). https://doi.org/10.1128/jvi.02304-20. https://journals.asm.org/doi/10.1128/JVI.02304-20.