Researchers have revealed the fine-scale genetic lineage structure of the coronavirus disease 2019 (COVID-19) epidemic in the UK earlier this year, as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first swept through the country.
By analyzing 50,887 SARS-CoV-2 genomes, the team has provided insights into the micro-epidemiological patterns underlying the features of one of the world's largest national COVID-19 epidemics.
The researchers from the UK and Canada say the exceptional size of their genomic survey enabled them to quantify the abundance, size distribution, and spatial range of SARS-CoV-2 transmission lineages across the UK during the first half of 2020.
Such information can provide a new context for the planning and evaluating future public health interventions, whether they be at the regional, national, or international scale.
A pre-print version of the paper is available in the server medRxiv*, while the article undergoes peer review.
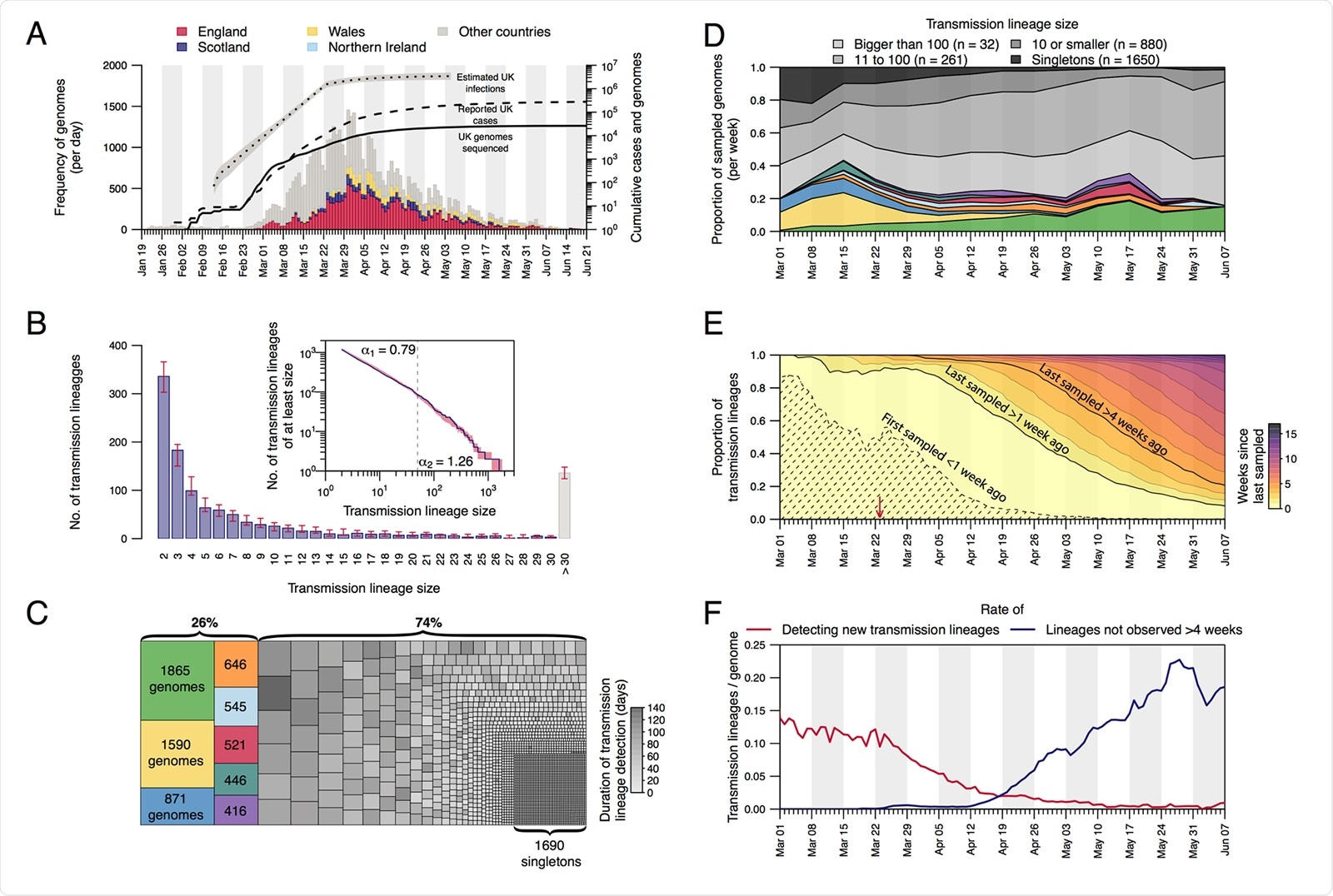
Structure and dynamics of UK transmission lineages. (A) Collection dates of the 50,887 genomes analyzed here (left-hand axis). Genomes are colored by sampling location (England=red, Scotland=dark blue, Wales=yellow, Northern Ireland=light blue, elsewhere=grey). The solid line shows the cumulative number of UK virus genomes (right-hand axis). The dashed and dotted lines show, respectively, the cumulative number of laboratory-confirmed UK cases (by specimen date) and the estimated number of UK infections (17; grey shading=95% CI; right-hand axis). Due to retrospective screening, the cumulative number of genomes early in the epidemic exceeds that of confirmed cases. (B) Distribution of UK transmission lineage sizes. Blue bars show the number of transmission lineages of each size (red bars=95% HPD of these sizes across the posterior tree distribution). Inset: the corresponding cumulative frequency distribution of lineage size (blue line), on double logarithmic axes (red shading=95% HPD of this distribution across the posterior tree distribution). Values either side of vertical dashed line show coefficients of power-law distributions (P[X ³ x] ~ xa) fitted to lineages containing ≤50 ( a 1) and >50 ( a 2) virus genomes, respectively. (C) Partition of 26,181 UK genomes into UK transmission lineages and singletons, colored by (i) lineage, for the 8 largest lineages, or (ii) duration of lineage detection (time between the lineage’s oldest and most recent genomes) for the remainder. (D) Lineage size breakdown of UK genomes collected each week. Colors of the 8 largest lineages are as depicted in (C). (E) Trends through time in the detection of UK transmission lineages. For each day, all lineages detected up to that day are colored by the time since the transmission lineage was last sampled. Isoclines correspond to weeks. Shaded area=transmission lineages that were first sampled <1 week ago. The red arrow indicates the start of the UK lockdown. (F) Red line=daily rate of detecting new transmission lineages. Blue line=rate at which lineages have not been observed for >4 weeks.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Viruses can be tracked using large-scale genome sequencing
Infectious disease epidemics are composed of multiple transmission chains, yet very little is understood about how co-circulating transmission lineages differ in size, persistence, and distribution. Similarly, little is understood about how the combined actions of these lineages contribute to important factors such as the size and duration of an epidemic.
However, recent studies of some viruses, including Ebola, Zika, and influenza, have shown that the emergence and spread of viruses can be tracked using large-scale genome sequencing.
These studies have demonstrated just how highly dynamic regional epidemics can be at the genetic level, with recurrent importation and extinction of transmission chains co-occurring within a given location, say Oliver Pybus (University of Oxford) and team.
"In addition to measuring genetic diversity, understanding pathogen lineage dynamics can help target interventions effectively, track variants with potentially different phenotypes, and improve the interpretation of incidence data," they add.
The COVID-19 epidemic experienced in the UK during early 2020 was one of the most extensive and most well-represented through genomic sampling.
The number of new SARS-CoV-2 infections increased during March, peaked in April, and by June 26th, 40,453 people had died from COVID-19.
What did the researchers do?
Pybus and colleagues combined an analysis of 50,887 SARS-CoV-2 genomes (including 26,181 from the UK during the first wave of infection) with epidemiological and travel data to characterize the genetic structure and lineage dynamics of the UK epidemic.
Prior to lockdown in the country, high volumes of travel and little restriction on arrivals from abroad had resulted in the establishment and co-circulation of more than 1,000 identifiable UK transmission lineages.
Lineages introduced before lockdown were larger and more dispersed
The eight largest lineages were first detected prior to the lockdown on March 23rd, and these larger lineages persisted for longer.
The detection of UK transmission lineages changed significantly over time. During early March, the epidemic was characterized by lineages that had first been observed within the previous week. By June 1st, on the other hand, more than 73% had not been detected for over 4 weeks, indicating that they were either rare or had become extinct.
These results suggest that the first epidemic wave resulted from the concurrent growth of many transmission lineages that had been introduced to the UK independently, says the team. They also suggest that the implementation of non-pharmaceutical interventions was followed by the extinction of lineages in a size-dependent manner.
Spatial distribution of UK transmission lineages
The study revealed that larger lineages were observed in more locations, indicating that they were more geographically widespread. With every additional 100 genomes in a transmission lineage, its range increased by 6 to 7 regions.
"These observations indicate substantial dissemination of a subset of lineages across the UK and suggest many regions experienced a series of introductions of new lineages from elsewhere, potentially hindering the impact of local interventions," say Pybus and colleagues.
Investigating introduction of the lineages
To investigate how transmission lineages were introduced to the UK, the team estimated the rate and source of SARS-CoV-2 importations into the country.
The researchers say importation was surprisingly dynamic, rising and falling dramatically over the course of just 4 weeks. Eighty percent of importations that gave rise to detectable transmission occurred between February 27th and March 30th.
Analysis of the country-specific contributions to virus importation showed that international arrivals' relative contributions were also highly dynamic.
Dominant source locations quickly changed during February and March, and the source locations became more diverse in mid-March.
"Earliest importations were most likely from China or elsewhere in Asia but were rare compared to those from Europe," say the researchers.
What are the implications of the study?
The team says that earlier lineages being larger, more dispersed, and more challenging to eliminate, emphasizes the importance of rapid or pre-emptive interventions in reducing transmission.
Although the UK lockdown coincided with importation restrictions and less regional lineage diversity, any resulting extinction of transmission lineages was size-dependent.
The over-dispersed nature of SARS-CoV-2 transmission probably exacerbated this effect, say Pybus and colleagues, thereby favoring longer survival of the larger, more widespread lineages and faster elimination of local ones in low-prevalence regions.
"The degree to which the surviving lineages contributed to the UK's ongoing second epidemic is currently under investigation," write the researchers.
"The transmission structure and dynamics measured here provide a new context in which future public health actions at regional, national, and international scales should be planned and evaluated," they add.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Pybus O, et al. Establishment & lineage dynamics of the SARS-CoV-2 epidemic in the UK. medRxiv, 2020. doi: https://doi.org/10.1101/2020.10.23.20218446, https://www.medrxiv.org/content/10.1101/2020.10.23.20218446v1
- Peer reviewed and published scientific report.
Plessis, Louis du, John T. McCrone, Alexander E. Zarebski, Verity Hill, Christopher Ruis, Bernardo Gutierrez, Jayna Raghwani, et al. 2021. “Establishment and Lineage Dynamics of the SARS-CoV-2 Epidemic in the UK.” Science (New York, N.Y.) 371 (6530): 708–12. https://doi.org/10.1126/science.abf2946. https://www.science.org/doi/10.1126/science.abf2946.