During the process of translation, cells use messenger RNA (mRNA) to synthesize proteins. Sometimes during translation, ribosomal or translational frameshifting can occur, which allows the production of several unique proteins from a single messenger RNA (mRNA). This has been mainly observed in viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus causing the COVID-19 pandemic.
In all coronaviruses, programmed -1 translational frameshifting, where the reading frame of the tri-nucleotide sequences is shifted back by one nucleotide, is conserved. This is necessary for the production of viral RNA-dependent RNA polymerase, RNA modification and processing, and RNA proofreading.
This frameshifting in SARS-related viruses occurs at the slippery sequence U_UUA_AAC, which is thought to form a 3-stemmed pseudoknot structure and change the reading frame UUU_AAA_C. Maintaining this frameshifting precisely is essential for viral infectivity. Mutation in even a single nucleotide in the frameshifting region of SARS-CoV-2 RNA prevents viral replication.
The frameshifting process does not occur in human cells, and this process in the SARS-CoV-2 virus could be a potential target for developing drugs.
The structure of a 3’ stimulatory pseudoknot in the viral genome or by itself has been proposed recently. Researchers report the structure and mechanisms of mammalian ribosomes during translation of the -1 frameshifting of the SARS-CoV-2 viral genome in a new study published on the preprint server bioRxiv*.
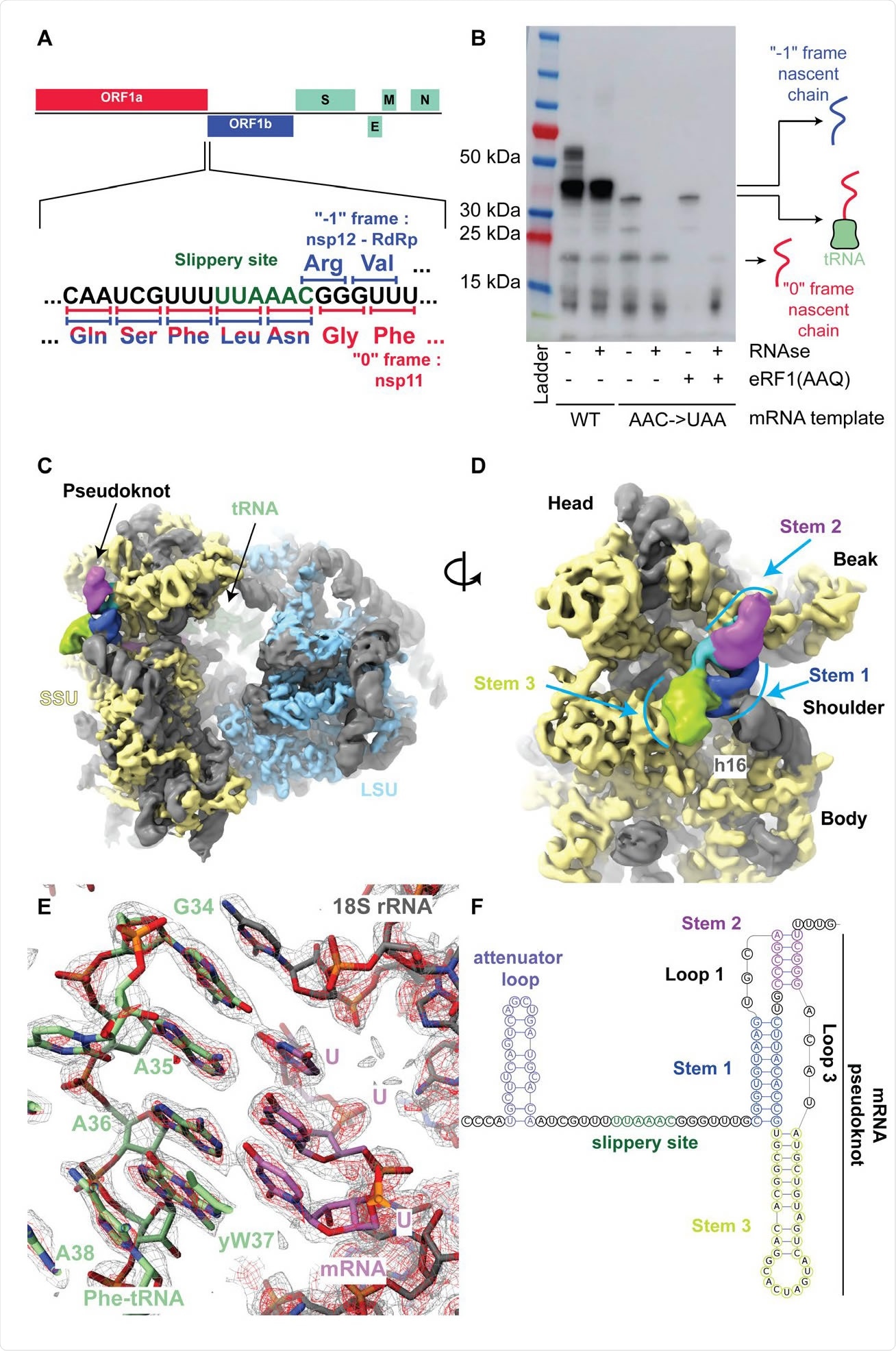
The SARS-Cov-2 pseudoknot interacts with the ribosome and pauses translation upstream of the slippery site. (A) Schematic of the SARS-CoV-2 main ORF. In the close up view of the frameshift event, codons and corresponding amino acids are shown. During -1 frameshifting, the ‘slippery site’ codons UUA (Leu) and AAC (Asn) are the last codons decoded in the 0 frame. Upon -1 frameshifting of the AAC codon to AAA, translation resumes at the CGG (Arg) triplet, where elongation proceeds uninterrupted to produce full-length Nsp12. (B) In vitro translation reaction depicting pausing at the frameshift site. Efficient frameshifting is observed for the WT template.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Visualizing ribosome during translation
The researchers from ETH Zurich, the University College Cork, and the University of Bern prepared a translation complex in vitro with a rabbit reticulocyte lysate system having viral mRNA with the slippery sequence U_UUA_AAC and another complex with a mutant mRNA that prevents -1 frameshifting.
Using cryo-electron microscopy, the scientists visualized the ribosomes in the process of translation at a 2.2-Å resolution to build an accurate structure of the mammalian 80S ribosome. They observed almost all the rRNA modification proposed for human ribosomes, as the modified residues are conserved in rabbit rRNA.
They also found a Phe-tRNA(Phe) bound to the P-site, suggesting the downstream pseudoknot pauses the ribosome so that the P-site tRNA interacts with the UUU codon just before the first codon of the frameshift site.
With a 5–7 Å resolution, the team was able to see how the RNA was folded in the pseudoknot area. The pseudoknot forms an H-Type structure, with Stems 1 and 2 stacked on top of each other and Stem 3 sticking out almost perpendicular to them.
The pseudoknot was lodged between the small ribosomal subunit's head and body, constraining any relative motions that would need to occur during translocation. In addition, the start of the pseudoknot was shifted by two nucleotides relative to the predicted structure.
Structures important in frameshifting
In SARS-CoV-2, the 0 frame stop codon is five codons from the frameshift site. The researchers found that increasing the distance of the 0 frame stop codon from the frameshift site led to decreased frameshifting efficiency. Removing it reduced frameshifting by almost half.
Thus, the stop codon plays a vital role in frameshifting efficiency. The authors suggest the stop codon allows the pseudoknot to refold without being restrained by the mRNA channel.
The authors saw the nascent chain corresponding to the viral polyprotein along the ribosomal exit tunnel during the paused translation. This chain interacts with the ribosomal tunnel, including at the constriction site, where arginine 4387 of Nsp10 interacts with the A1555 of 28S rRNA and is stabilized by leucine 4386. The C-terminal end of Nsp10 shows a partially folded zinc finger motif further into the tunnel.
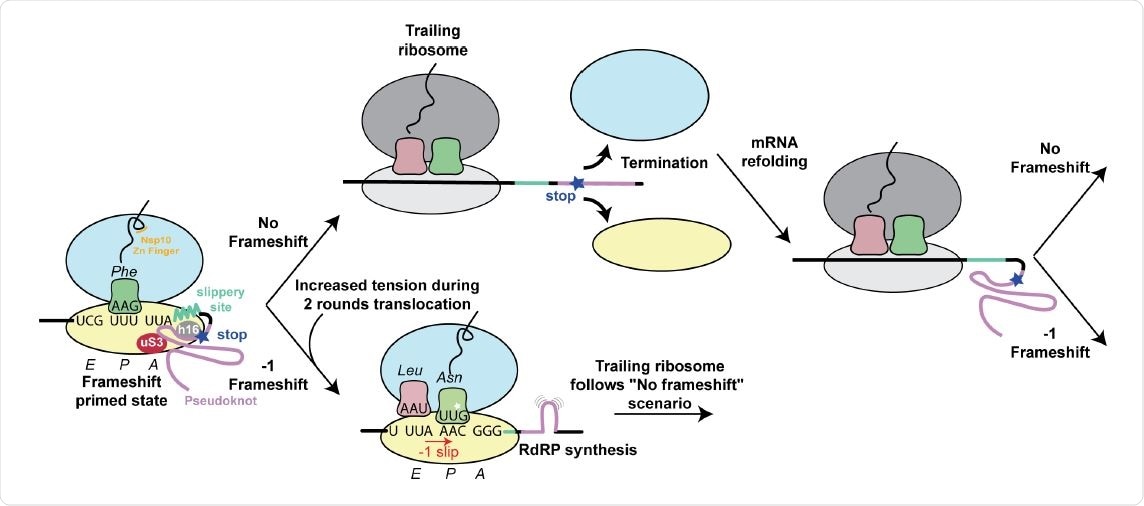
Structure based model for -1 programmed frameshifting in coronaviruses and its regulation. The observed interactions between the pseudoknot and the ribosome prime the system for frameshifting. The features of the pseudoknot and the interactions between the nascent chain and the ribosomal tunnel play a role in the efficiency of frameshifting. The efficiency of frameshifting is increased by the presence of a stop codon near the frameshifting site. Ribosomes that progress beyond the frameshifting site in the 0 frame quickly terminate and disassemble, thereby increasing the chances that the pseudoknot will refold before it is encountered by the closely trailing ribosome. The trailing ribosome in turn encounters the pseudoknot, which increases the possibility of undergoing -1 frameshifting.
When the researchers used an in vitro assay to understand the nascent chain's effect on the frameshifting efficiency, they found a 30% increase in frameshifting when the leucine 4386 and arginine 4387 were replaced with alanine. This suggests nascent chain interactions with the ribosomal exit tunnel are important in modulating frameshifting levels.
Thus, nascent-chain factors, or even intracellular zinc concentration, which changes with viral infection, could influence frameshifting rates.
Disrupting the frameshifting process may prevent SARS-CoV-2 from replicating. The authors demonstrated this was true by using a compound described before that inhibits frameshifting, and they found reduced virus levels in infected African green monkey Vero cells.
Thus, interfering with the interactions between the pseudoknot and the mRNA channel or during the initial chain interactions with the ribosomal tunnel to form the frameshift stimulatory pseudoknot could be potential drug development strategies
References and Further Reading

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bhatt, P. R. et al. (2020) Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. bioRxiv. https://doi.org/10.1101/2020.10.26.355099, https://www.biorxiv.org/content/10.1101/2020.10.26.355099v1
- Peer reviewed and published scientific report.
Bhatt, Pramod R., Alain Scaiola, Gary Loughran, Marc Leibundgut, Annika Kratzel, Romane Meurs, René Dreos, et al. 2021. “Structural Basis of Ribosomal Frameshifting during Translation of the SARS-CoV-2 RNA Genome.” Science, May. https://doi.org/10.1126/science.abf3546. https://www.science.org/doi/10.1126/science.abf3546.