By exploiting type 1 interferon's ability to foster an antagonistic cellular environment for viral replication, a research group from France pinpointed DEAD-box RNA helicase DDX42 as an intrinsic inhibitor of HIV, but also other pathogenic viruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Chikungunya virus. This compelling study is currently available on the bioRxiv* preprint server.
During the last 20 years, a myriad of cellular proteins with various functions have been identified as capable of hindering different steps in the HIV life cycle. This effect is particularly strong when human cells are predisposed to interferon.
As expected, the quest was accelerated with the introduction of genome-wide CRISPR/Cas9 knock-out genetic screens, which are touted as extremely steadfast approaches to unveil new regulators of viral infections.
Many RNA helicases from the DEAD box family (found in almost all organisms where they play pivotal roles in RNA metabolism) are well-known to control the HIV life cycle. Some notable examples are DDX3, DDX6, and DDX17; however, the impact of DDX42 on HIV replication had never been studied.
This specific helicase grabbed the attention of a group of scientists from the Université de Montpellier and Montpellier GenomiX in France. They aimed to appraise the impact of endogenous DDX42 on HIV-1 infection, as well as the consequences of enzyme overexpression.
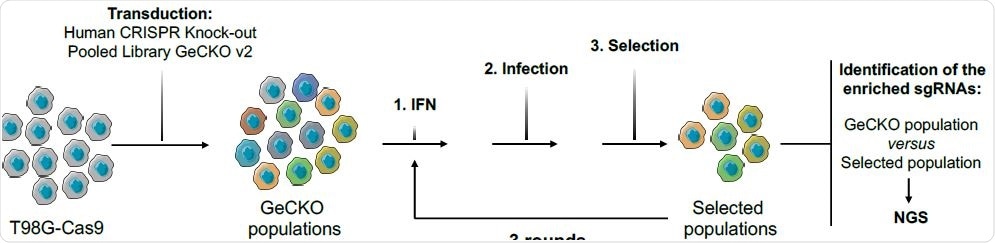
Methods and rationale

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The researchers used the Genome-Scale CRISPR Knock-Out (GeCKO) single guide RNA library in order to generate cell populations knocked-out for almost every human gene. Furthermore, a model T98G glioblastoma cell line was used since it is highly permissive to lentiviral infection and also able to suppress HIV infection following interferon treatment.
DDX42 is a known non-processive helicase, which also has RNA annealing activities and the propensity to displace RNA-binding proteins from single-stranded RNAs. In order to confirm DDX42's specific effect on HIV-1 infection, the researchers used three different small interfering RNAs to knockdown DDX42 expression.
The impact of DDX42 depletion was not tested only for retroviruses and retroelements, but also for five other viral families: the orthomyxovirus influenza A virus, the alphavirus chikungunya virus, the flavivirus Zika virus, the rhabdovirus vesicular stomatitis virus, as well as SARS-CoV-2 responsible for the ongoing coronavirus disease (COVID-19) pandemic.
Broad antiviral activity of endogenous DDX42
"Here, we identified for the first time the DDX42 RNA helicase as an intrinsic inhibitor of HIV-1, capable of limiting the efficiency of viral DNA accumulation", say study authors in this exciting bioRxiv paper.
"More specifically, our study revealed broad activity of endogenous DDX42 among retroviruses and retroelements, which was observed in various cell types – including primary CD4+ T cells", they add.
The depletion of endogenous DDX42 increased the infection with HIV – both in model cell lines and T cells or monocyte-derived macrophages (which are physiological targets of HIV). This was observed regardless of the interferon treatment.
Likewise, overexpressing the dominant-negative mutant of DDX42 had a positive impact on HIV infection, while wild-type DDX42 overexpression had a potent inhibitory effect. Moreover, an endogenous DDX42 depletion was directly linked to an increase in viral DNA accumulation.
An interesting finding was that DDX42 was able to inhibit viruses belonging to other families, with quite different replication strategies, which includes SARS-CoV-2 and chikungunya. Nonetheless, DDX42 did not show an impact on all the tested viruses, akin to some other broad-spectrum antiviral inhibitors such as MxA proteins.
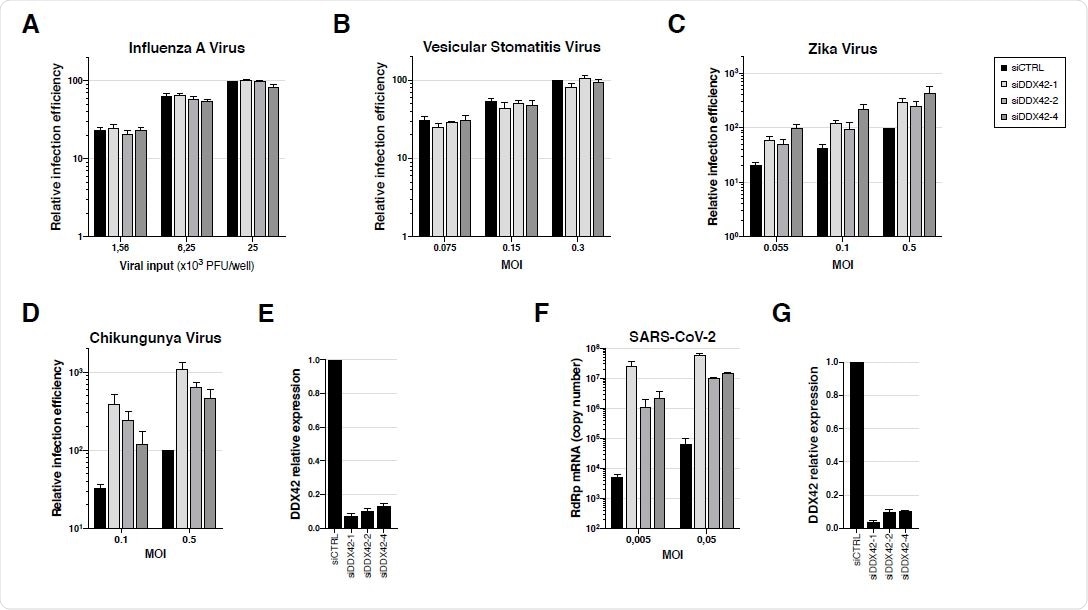
Endogenous DDX42 does not impact VSV or IAV infection, is a mild inhibitor of ZIKV infection and potently inhibits both CHIKV and SARS-CoV-2 replication. A. U87-MG cells were transfected with a nontargeting siRNA or siRNAs targeting DDX42 (siDDX42-1, -2 and -4) and, 72 h post-transfection, cells were infected with an influenza A reporter virus expressing Nanoluciferase. The Nanoluciferase signals were measured 16 h later. B. Control- and DDX42-depleted U87-MG cells were infected with a Firefly expressing VSV reporter at the indicated MOIs. The Firefly signals were measured 24 h later. C. Control- and DDX42-depleted U87-MG cells were infected with a Nanoluciferase-expressing ZIKV at the indicated MOIs. The Nanoluciferase signals were measured 24 h later. D. Control- and DDX42-depleted U87-MG cells were infected with a Gaussiaexpressing CHIKV at the indicated MOIs. The Gaussia signals were measured 24 h later. E. Quantification of DDX42 silencing efficiency in U87-MG cells by RT-qPCR. F. A549-ACE2 cells were transfected with a nontargeting siRNA or siRNAs targeting DDX42 (siDDX42-1, -2 and -4). 72 h post-transfection, cells were infected with SARS-CoV-2 virus at the indicated MOIs and lysed 2 days later, their RNA contents extracted and SARSCoV- 2 replication efficiency was measured by RT-qPCR, using RdRp primers and probe. G. Quantification of DDX42 silencing efficiency in A549-ACE2 cells by RT-qPCR. A-G Data represent the mean ± S.E.M. of three independent experiments.
Direct alteration of viral ribonucleoproteins
By observing a close proximity between DDX42 and HIV capsid, a direct mode of action on viral ribonucleoprotein (RNP) complexes could be hypothesized, which is also backed by the available scientific literature.
"Nonetheless, further investigation will be needed to determine whether DDX42 acts directly by altering viral ribonucleoproteins, and, if that's the case, what are the determinants for viral RNP recognition", caution study authors.
In conclusion, this study underscores the importance of understanding the exact mechanisms of action of DDX42 RNA helicase, as well as its contribution in controlling replication of RNA viruses. This may in turn inform future antiviral intervention strategies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bonaventure, B. et al. (2020). A genome-wide CRISPR/Cas9 knock-out screen identifies the DEAD box RNA helicase DDX42 as a broad antiviral inhibitor. bioRxiv. https://doi.org/10.1101/2020.10.28.359356, https://www.biorxiv.org/content/10.1101/2020.10.28.359356v1
- Peer reviewed and published scientific report.
Bonaventure, Boris, Antoine Rebendenne, Ana Luiza Chaves Valadão, Mary Arnaud‐Arnould, Ségolène Gracias, Francisco Garcia de Gracia, Joe McKellar, et al. 2022. “The DEAD Box RNA Helicase DDX42 Is an Intrinsic Inhibitor of Positive‐Strand RNA Viruses.” EMBO Reports 23 (11). https://doi.org/10.15252/embr.202154061. https://www.embopress.org/doi/full/10.15252/embr.202154061.