The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has already infected more than 46.83 million people globally and claimed over 1.2 million lives so far. The molecular pathophysiology behind the clinical manifestations of SARS-CoV-2 infection and disease course of COVID-19 is complex and encompasses multiple cell types, pathways, and organs.
In order to gain insights into this complex network, there is a need for the biomedical research community to approach it from a systems perspective, collect knowledge from all available scientific literature and databases, and integrate the information using biology standards. With this in mind, a collaboration of researchers from 120 different institutes across 30 countries including France, Germany, Australia, Japan, USA, India, Turkey, Brazil, and Canada initiated an effort to develop a COVID-19 Disease Map - a collection of computational models and diagrams of molecular mechanisms implicated in COVID-19.
An unprecedented community-driven biocuration of published data on COVID-19 pathophysiology
This map is a large-scale community effort to develop an open-access and computable repository of molecular mechanisms linked to COVID-19 pathophysiology. The multi-faceted team comprised more than 230 biocurators, modelers, domain experts, bioinformaticians, computational biologists, and data analysts.
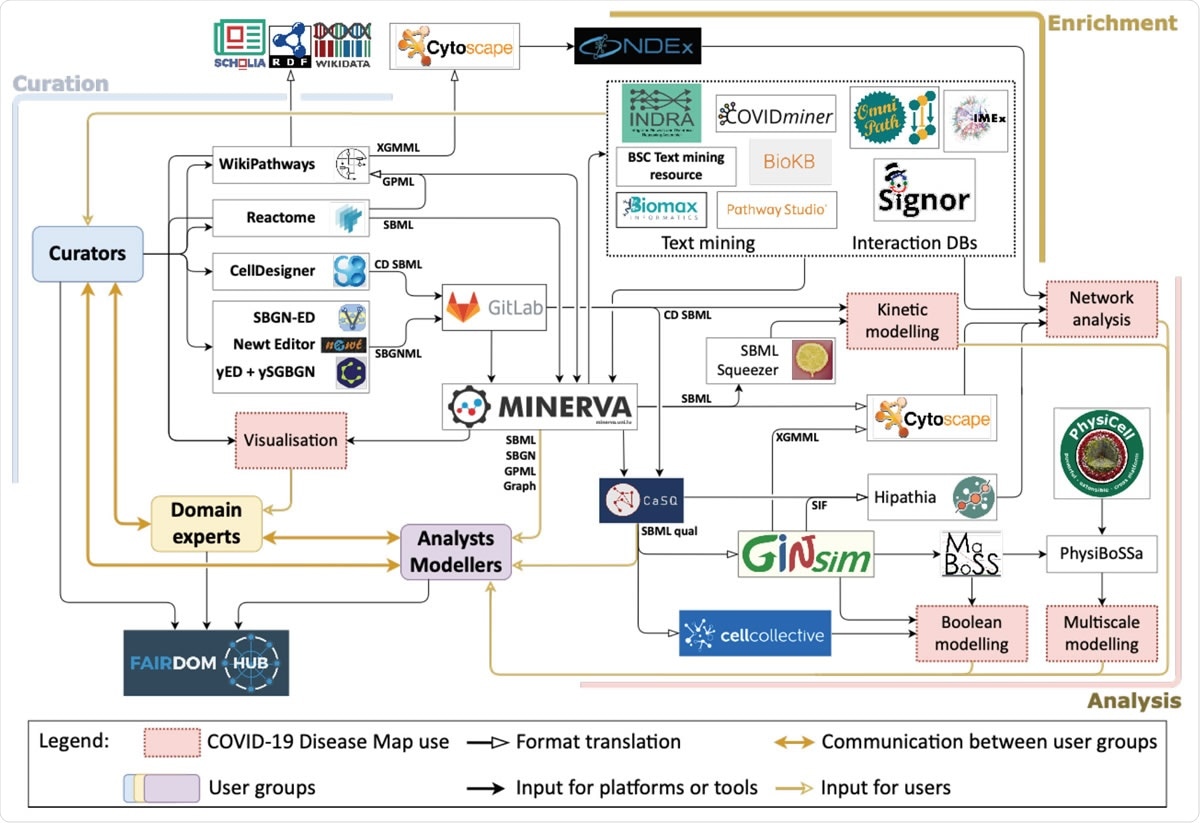
The ecosystem of the COVID-19 Disease Map Community.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Their work, published on the preprint server bioRxiv*, discusses the tools, guidelines, and platforms required to develop this comprehensive disease map and highlight the role played by the databases and text mining approaches in the enrichment validation of the curated information. The researchers also describe the contents of the COVID-19 map and how relevant they are to disease's molecular pathophysiology. They explore analytical and computational modeling approaches that can be applied to the COVID-19 Disease Map for mechanistic data interpretation and predictions and also demonstrate robust applications of this map in several scenarios.
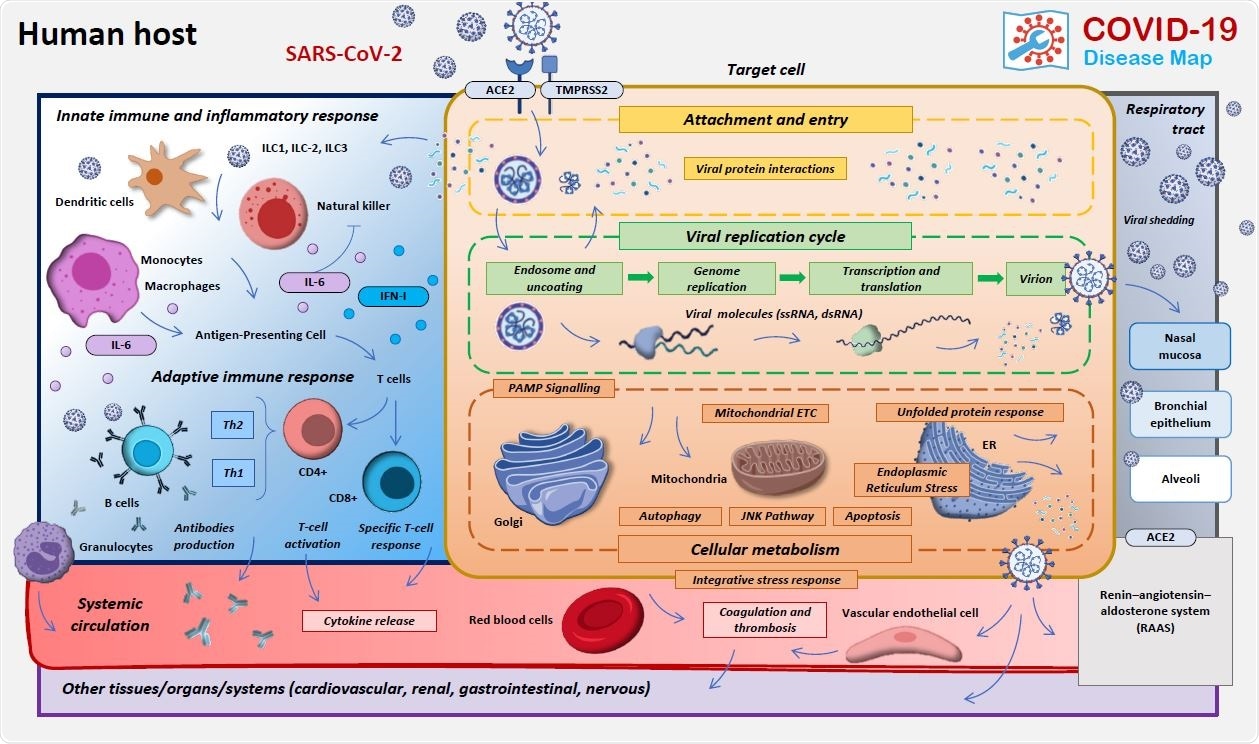
The structure and content of the COVID-19 Disease Map. The areas of focus of the COVID-19 Map biocuration.
COVID-19 Disease Map - a shared mental map of the dynamic nature of COVID-19 at the molecular and systemic levels
While the COVID-19 Disease Map is a knowledgebase and computational repository, it is also a graphical and interactive representation of molecular mechanisms relevant to the disease, which links many knowledge sources.
As a computational resource, it offers curated content for graph-based analyses and disease modeling. It acts as a shared mental map to better understand the dynamic nature of COVID-19 at the molecular level and its propagation at a systemic level.
Thus, the map serves as a platform for precise model formulations, accurate interpretation of data, therapy monitoring, and exploring drug repositioning potential. It spans three platforms and assembles diagrams on COVID-19 molecular mechanisms.
These diagrams are chosen from relevant, published SARS-CoV-2 studies and completed where necessary using mechanisms discovered in other viruses of the same family. This unprecedented effort in terms of community-driven biocuration resulted in more than 40 diagrams with molecular resolution built since March 2020.
"With new results published every day, and with the active engagement of the research community, we envision the COVID-19 Disease Map as an evolving and continuously updated knowledge base whose utility spans the entire research and development spectrum from basic science to pharmaceutical development and personalized medicine."
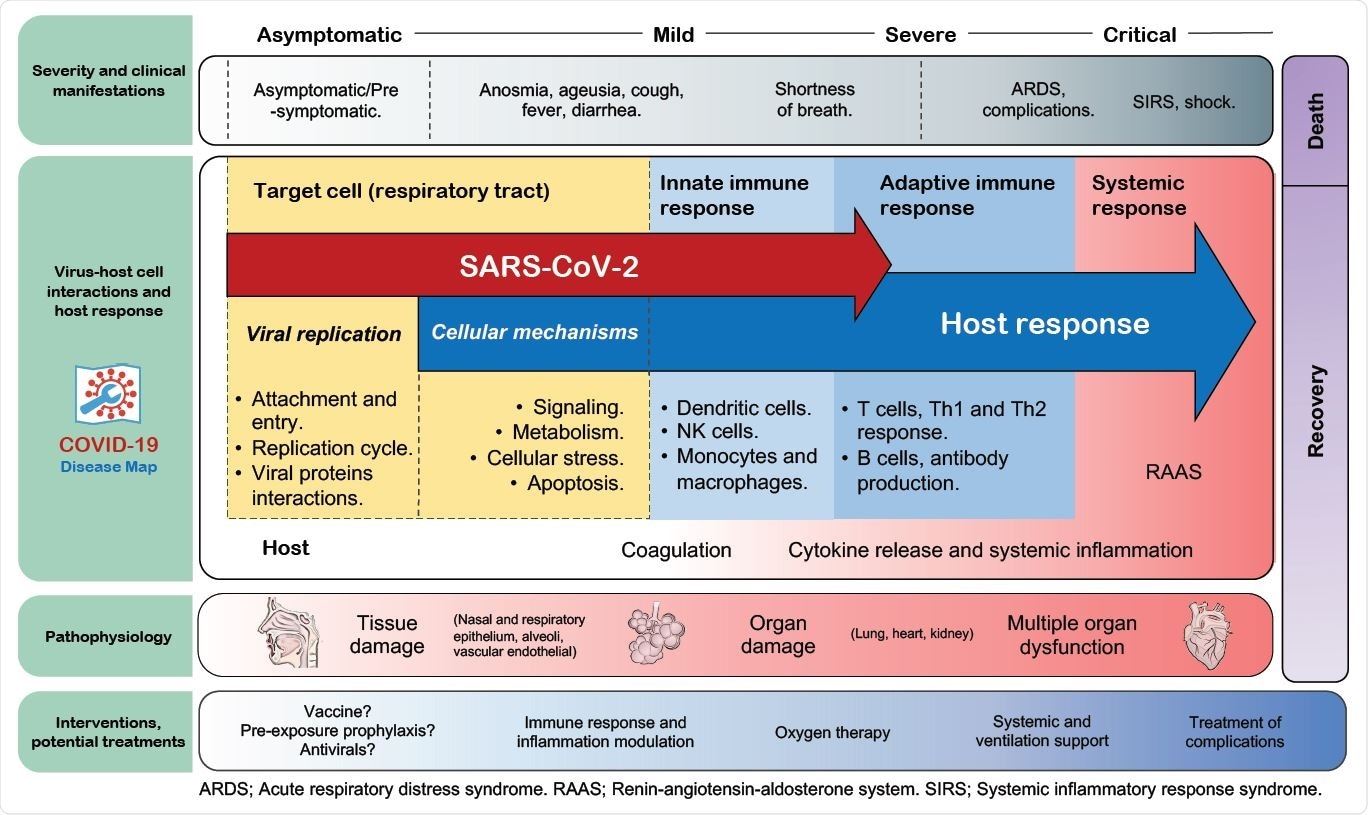
Overview of the map in the context of COVID-19 progression. Pathways and cell types involved in the sequential stages of COVID-19, including some of the most common clinical manifestations and medical management from the moment of infection to the disease resolution, are shown. The distribution of the elements is for illustrative reference and does not necessarily indicate either a unique/static interplay of these elements or an unvarying progression.
COVID-19 Disease Map - a tipping point to easy data generation and knowledge accumulation
According to the authors, this shows that biocuration expertise, clear guidelines, and text mining solutions can speed up the evolution of published findings into a meaningful representation of knowledge. The authors believe that the COVID-19 Disease Map can be a tipping point to easy research data generation and knowledge accumulation. Their approach leveraged a massive bioinformatics community's capacity and expertise and brought them together to enhance the way the scientific community builds and shares knowledge. By aligning their efforts, they synchronize relevant content with similar resources, provide COVID-19 specific pathway models, and promote discussion and feedback at all curation stages.
Their approach includes a large-scale effort to produce interoperable tools and seamless downstream analysis pipelines to improve the applicability of existing methodologies to the COVID-19 Disease Map content. With the help of this map, the researchers aim to build armor for new treatments that will potentially prevent new waves of COVID-19 or similar future pandemics.
"We aim to provide the tools to deepen our understanding of the mechanisms driving the infection and help boost drug development supported with testable suggestions."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Marek Ostaszewski et al, COVID-19 Disease Map, a computational knowledge repository of SARS-CoV-2 virus-host interaction mechanisms, bioRxiv 2020.10.26.356014; doi: https://doi.org/10.1101/2020.10.26.356014, https://www.biorxiv.org/content/10.1101/2020.10.26.356014v1
- Peer reviewed and published scientific report.
Ostaszewski, Marek, Anna Niarakis, Alexander Mazein, Inna Kuperstein, Robert Phair, Aurelio Orta‐Resendiz, Vidisha Singh, et al. 2021. “COVID19 Disease Map, a Computational Knowledge Repository of Virus–Host Interaction Mechanisms.” Molecular Systems Biology 17 (10). https://doi.org/10.15252/msb.202110387. https://www.embopress.org/doi/full/10.15252/msb.202110387.