Amidst the frantic global search for a vaccine to inhibit SARS-CoV-2 or a drug to treat coronavirus 2019 (COVID-19), remdesivir has been recently approved by the U.S. FDA to treat COVID-19 patients.
The causative agent of COVID-19, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has exhibited a positive response to this nucleoside analog drug.
The virus uses an RNA-dependent RNA polymerase (RdRp) to replicate. The active form of remdesivir inhibits the RdRp and exhibits antiviral activity. However, the specific mechanism by which remdesivir inhibits the RdRp is not completely clear.
In a recent bioRxiv* preprint, a research team from Germany, Goran Kokic et al. study the molecular mechanism of remdesivir-induced stalling of RdRp - using synthetic RNA chemistry, biochemistry, and cryo-electron microscopy.
Remdesivir is a phosphoramidite prodrug that is metabolized in cells to yield an active nucleoside triphosphate (NTP) analog, referred to as remdesivir triphosphate (RTP) in this study. RdRp can use RTP as a substrate, leading to the incorporation of remdesivir monophosphate (RMP) into the growing RNA. The previous studies show how RMP is incorporated into the RNA. However, how the drug remdesivir inhibits the RdRp is explored here. Unique to coronaviruses, RdRp stalling occurs only after three more nucleotides have been added to the RNA.
In this study, the researchers investigate the following: 1) how the presence of the drug (RTP) influences the polymerase action of elongation, 2) determine the structures of RdRpRNA complexes, 3) trap the states of the RdRp complex structurally, that are relevant for understanding remdesivir-induced RdRp stalling, 4) does the RdRp stalling results from a translocation barrier.
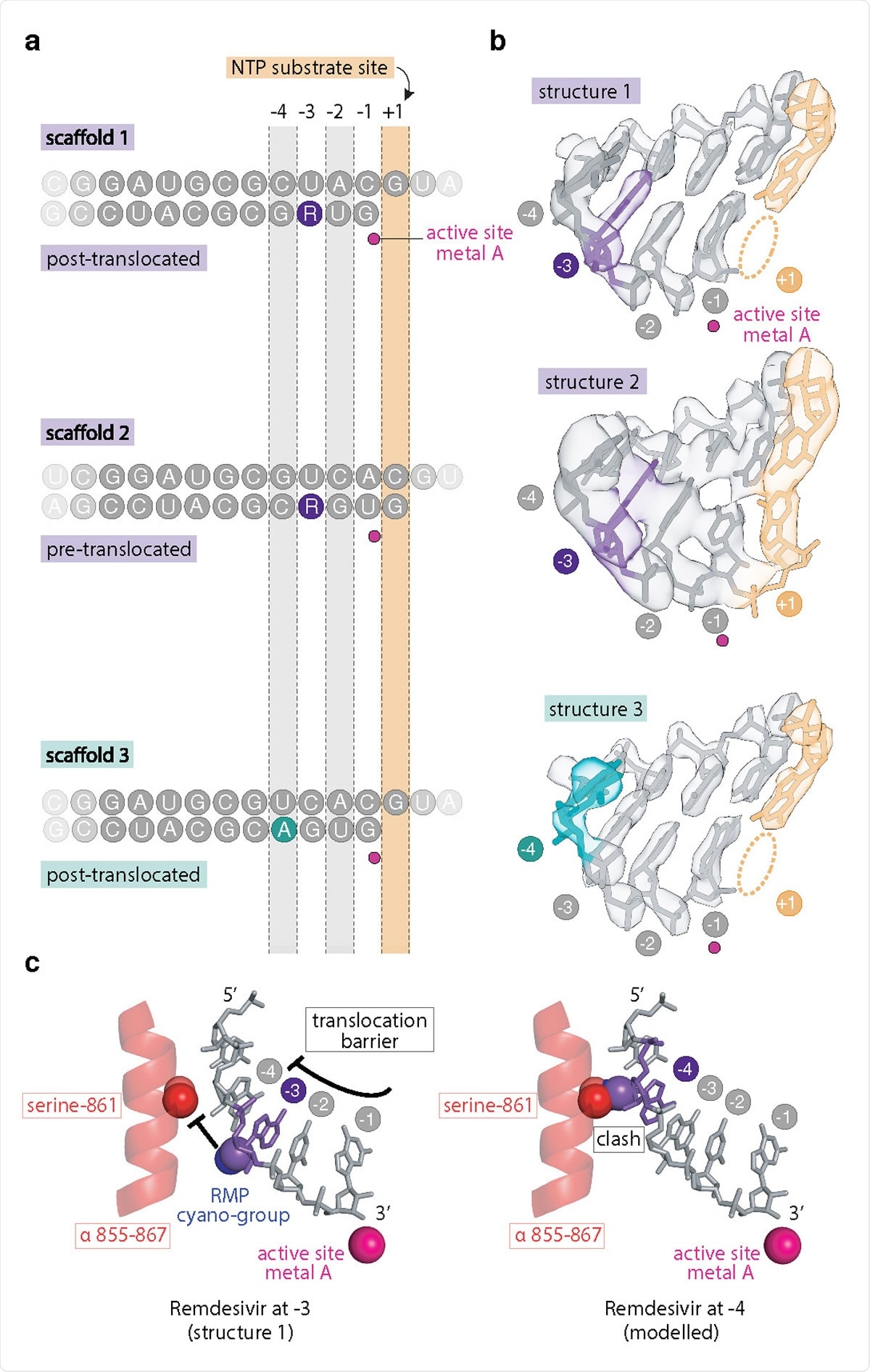
Structural analysis of remdesivir-induced RdRp stalling. a Position of RNA scaffolds 1–3 as observed in RdRp-RNA complex structures 1–3. Template and product strands are on the top and bottom, respectively. b Cryo-EM density of RNA in the active center of structures 1–3. The active site metal ion was modelled30 and is shown as a magenta sphere. c The C1’-cyano group of the RMP ribose moiety (violet) is accommodated at position –3 (left), but would clash with the side chain of nsp12 residue serine-861 (red) at position –4 (right). Spheres indicate atomic van der Waals surfaces.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
RdRp uses RTP as a substrate, leading to the incorporation of RMP into a growing RNA product extending by three more nucleotides before it stalls. This stalling mechanism is specific to coronaviruses. For example, in the case of the Ebola virus, RdRp adds five RNA nucleotides after RMP incorporation before it stalls.
The researchers confirmed the presence of RMP in the obtained RNA oligonucleotides using denaturing HPLC and LC-MS. They also confirmed that the presence of RMP inhibits RNA extension by RdRp on minimal RNA template-product scaffolds.
Post RMP incorporation, the researchers investigated RdRp-RNA complexes - after the addition of two or three nucleotides (using cryo-EM analysis). The RdRp-RNA structures resolution is determined, also revealing the positions of RMP in the RNA product strand.
This study shows that the fourth nucleotide's addition is impaired by a barrier to further the RNA translocation. This causes retention of the RNA 3'-nucleotide in the active site of the RdRp, not allowing the next nucleoside triphosphate, thereby stalling the enzyme.
The researchers found that the translocation barrier results from the sterically impaired passage of the cyano group in RMP past the serine-861 side chain in the nsp12 (nonstructural) subunit of RdRp.
These results are consistent with biochemical studies and also previous data.
In this study, the researchers show how the remdesivir-stalled state escapes viral proofreading. The matched RNA 3'-end may escape proofreading because the viral exonuclease nsp14 preferentially recognizes a mismatched 3'-end.
However, the researchers also point out that some proofreading may occur, rendering remdesivir less efficient (the viral exonuclease can remove several nucleotides from the base-paired RNA 3'-end).
It is important to note that at high NTP concentrations, RdRp stalling was overcome, resulting in the formation of the full-length RNA; despite the presence of RMP in the RNA product. Because of these results, the researchers note that remdesivir neither acts as a chain terminator nor as an elongation block but rather triggers delayed RdRp stalling by an unknown mechanism.
This paper's mechanistic insights are critical to understanding the inhibition process of an approved antiviral drug used against SARS-CoV-2 and may further the search for compounds with improved potential in this direction to interfere with coronavirus replication.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mechanism of SARS-CoV-2 polymerase inhibition by remdesivir Goran Kokic, Hauke S. Hillen, Dimitry Tegunov, Christian Dienemann, Florian Seitz, Jana Schmitzova, Lucas Farnung, Aaron Siewert, Claudia Höbartner, Patrick Cramer bioRxiv 2020.10.28.358481; doi: https://doi.org/10.1101/2020.10.28.358481, https://www.biorxiv.org/content/10.1101/2020.10.28.358481v1
- Peer reviewed and published scientific report.
Kokic, Goran, Hauke S. Hillen, Dimitry Tegunov, Christian Dienemann, Florian Seitz, Jana Schmitzova, Lucas Farnung, Aaron Siewert, Claudia Höbartner, and Patrick Cramer. 2021. “Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir.” Nature Communications 12 (1). https://doi.org/10.1038/s41467-020-20542-0. https://www.nature.com/articles/s41467-020-20542-0.