The initial cases of the coronavirus disease 2019 (COVID-19) were first reported in March 2020 in South Africa. Since then, the disease has spread rapidly and widely throughout the region. Unique mutations leading to 16 new lineages of the causal agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), have been identified in the South African population.
Transmission electron micrograph of SARS-CoV-2 virus particles, isolated from a patient. Image captured and color-enhanced at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Published in a recent medRxiv* paper, researchers from South Africa, the United Kingdom, and Brazil found that these mutations in SARS-CoV-2 lineages are localized to South Africa. These unique strains, not found elsewhere in the world, are thought to have contributed around 42% to the country's total infection rate.
The SARS-CoV-2 is a novel beta coronavirus, first detected in Wuhan, China, in late December 2019. In March 2020, the World Health Organization (WHO) declared the virus a global pandemic, and, with the rapid spread of the COVID-19 across the world since the end of last year, there have been over 47.3 million cases confirmed worldwide. Mitigation strategies such as lockdown, travel restrictions, and closure of non-essential businesses and schools have varied from country to country, and these have dictated the profile of local epidemic outbreaks. This illuminating study of South Africa's unique mutations of the virus unveils the sorts of effects these mitigatory measures can have on viral mutations and lineage behavior.
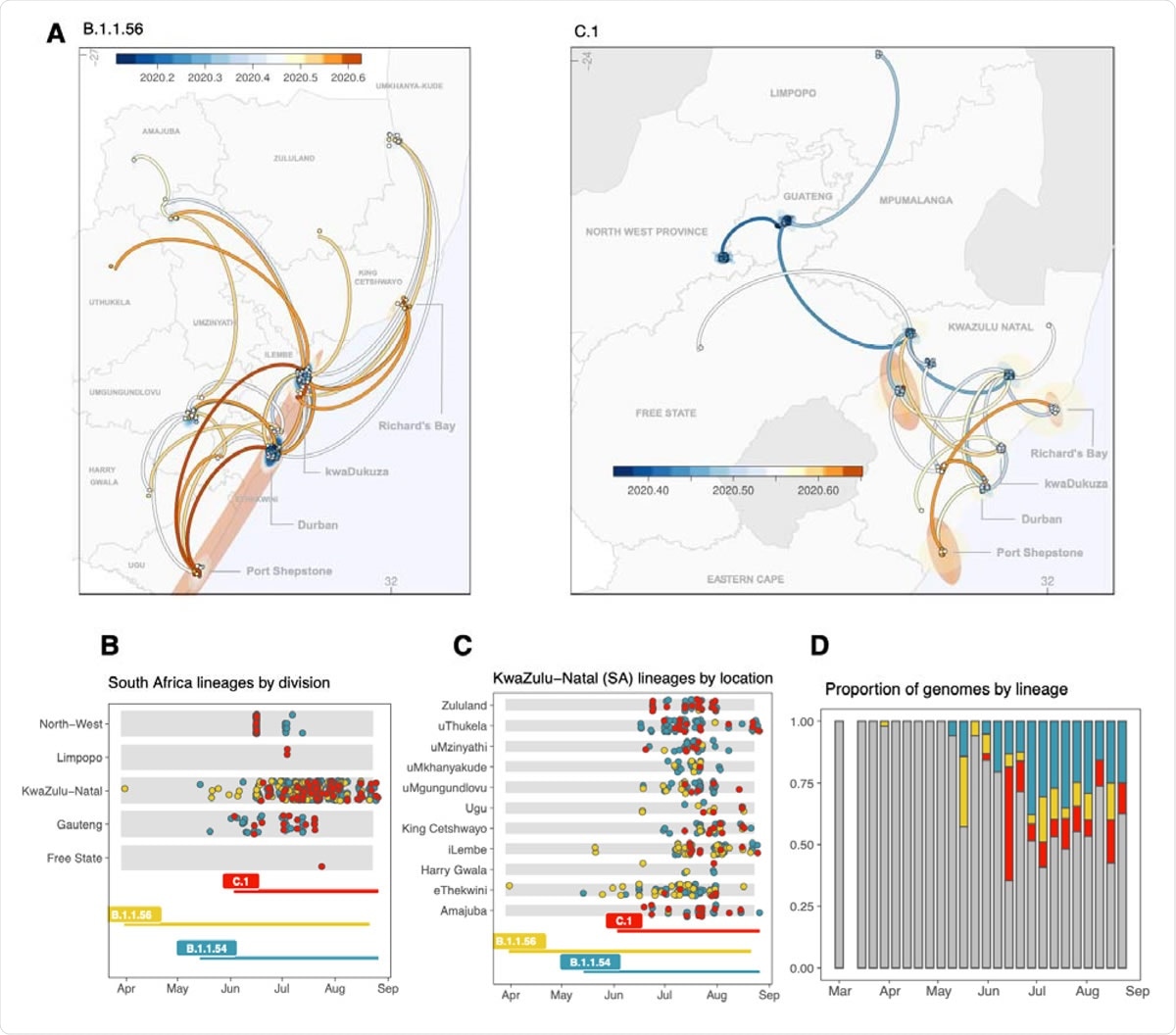
Geographical distribution and spread of lineage clusters in (5) five provinces and all districts of KZN. A) Mapping the spread of the B.1.1.56 cluster (left) and the C.1 cluster (right) from phylogeographic reconstructions. Time scale is specified in decimal dates from 2020.2 (March 2020) to 2020.6 (July 2020). Eastern Cape misspelled – to be fixed in next iteration. B) Sampling timeline and locations of genomes belonging to each lineage cluster in (5) five provinces. C) Sampling timeline and locations of genomes belonging to each lineage cluster in all 11 districts of KZN province. D) The progression of the proportions of genomes belonging to the main lineage clusters over time (B.1.1.54: blue, B.1.1.56: yellow, C.1: red), grey: others).
The study also charts the specific lineages' behavior, and such genomic epidemiology investigations help understand the emerging virus behavior better. SARS-CoV-2 has typically been classified into two main phylogenetic lineages: lineage A and lineage B - both originating in China. The first South African lineage to be identified is the B.1.106 lineage, and the most geographically widespread lineage in South Africa is the C1 lineage.
In this study, the first identified C lineage of SARS-CoV-2, C.1, has 16 mutations when compared to the original Wuhan sequence. The study also found that C1 has caused infections in multiple provinces, with the KwaZulu-Natal (KZN) as the most sampled province.
To understand the spread of the SARS-CoV-2, the research team formed the Network for Genomics Surveillance in South Africa (NGS-SA) - a consortium of genomics and bioinformatics scientists that work with national government laboratories to generate and analyze data in the country quickly.
They analyzed 1,365 high-quality whole genomes and mapped the emergence of 16 South African specific novel lineages. Among these established lineages, three lineages mainly caused over half of the infections in South Africa. The sampling was consistent from the beginning of the epidemic and corresponded to important events in the virus's epidemiological progression.
This interdisciplinary research team reports an in-depth analysis of the spread of SARS-CoV-2 in South Africa; they show that the bulk of introductions happened before lockdown and travel restrictions were implemented at the end of March 2020. Therefore, despite the restriction measures, the virus spread rapidly, causing more than 600,000 confirmed cases of SARS-CoV-2 infection (as per the period when the study was conducted: before September 2020).
The team also studied the nosocomial outbreaks of the infection: the B.1.106 lineage behavior was studied in a large hospital in the country. The active outbreak investigation itself may have limited the spread of this lineage. The phylogenetic tree of B.1.106 and its dispersion throughout KwaZulu-Natal (KZN) is presented here. This lineage is responsible for 16% of the infections in KZN at the start of April. However, with strict lockdown measures, it was controlled.
It is currently unknown if the newly mutated South African SARS-CoV-2 lineages have any advantage - in terms of transmission, viral replication, or reduced immunogenicity in the South African population - over older strains of the virus originating in Wuhan, China. However, the infectivity among the different viral strains does not differ even though 1) their mutations are synonymous and 2) the Ct values do not differ appreciably.
This study shows that several SARS-CoV-2 lineages, each with unique mutations, have emerged within localized epidemics during lockdown; even when mitigation methods were curbing the entry of new lineages from outside South Africa.
The researchers emphasize the usefulness of integrating genomic surveillance methods to understand SARS-CoV-2 spread in local settings. The near real-time genomic monitoring and data inference help in developing effective strategies to control the infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tegally, Houriiyah, et al. Major new lineages of SARS-CoV-2 emerge and spread in South Africa during lockdown. (October, 2020), medRxiv 2020.10.28.20221143; doi: https://doi.org/10.1101/2020.10.28.20221143, https://www.medrxiv.org/content/10.1101/2020.10.28.20221143v1
- Peer reviewed and published scientific report.
Tegally, H., Wilkinson, E., Lessells, R. J., Giandhari, J., Pillay, S., Msomi, N., Mlisana, K., Bhiman, J. N., von Gottberg, A., Walaza, S., Fonseca, V., Allam, M., Ismail, A., Glass, A. J., Engelbrecht, S., Van Zyl, G., Preiser, W., Williamson, C., Petruccione, F., & Sigal, A. (2021). Sixteen novel lineages of SARS-CoV-2 in South Africa. Nature Medicine, 27(3), 440–446. https://doi.org/10.1038/s41591-021-01255-3, https://www.nature.com/articles/s41591-021-01255-3