Using dynamic contact maps and energy differences between different conformations of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein (or S-protein), an international team of researchers have recently published an article on the bioRxiv* preprint server showing the different stable conformations of the spike protein. Determining potential target sites for destabilizing the spike protein can help develop drugs inhibiting its binding to host cells.
The spread of the coronavirus disease 19 (COVID-19) has researchers racing to understand how the SARS-CoV-2 pathogen, which causes the disease, infects and replicates in host cells.
It is established that the virus's spike protein is key in viral entry into the host cell. It binds to the human angiotensin-converting enzyme 2 (ACE2) and undergoes many conformational changes. The protein changes from the down to up receptor-binding domain (RBD) conformation, which prepares it for binding to ACE2, and then a subsequent fusion of the viral membrane with the host cell membrane.
Several studies have investigated how the changes in virus protein conformations allow it to infect host cells so easily. Computational studies show a correlation between the polybasic furin cleavage site and surface residues in the RBD region that recognizes ACE2, mediated by electrostatic interactions 10 nm apart. In the dominant virus mutation D614G, residues correlated with the mutation are located 7–10 nm from the SARS-CoV-2 RBD.
It has been suggested these changes could increase its transmissibility and aid the steps after fusion with host cells. However, since the spike protein region is generally unstable without the presence of the ACE2 receptor, experimentally studying the conformation is difficult.
A previous modeling study has shown the presence of a dynamic asymmetry that triggers a change in conformation, with the closed conformation being the ground state.
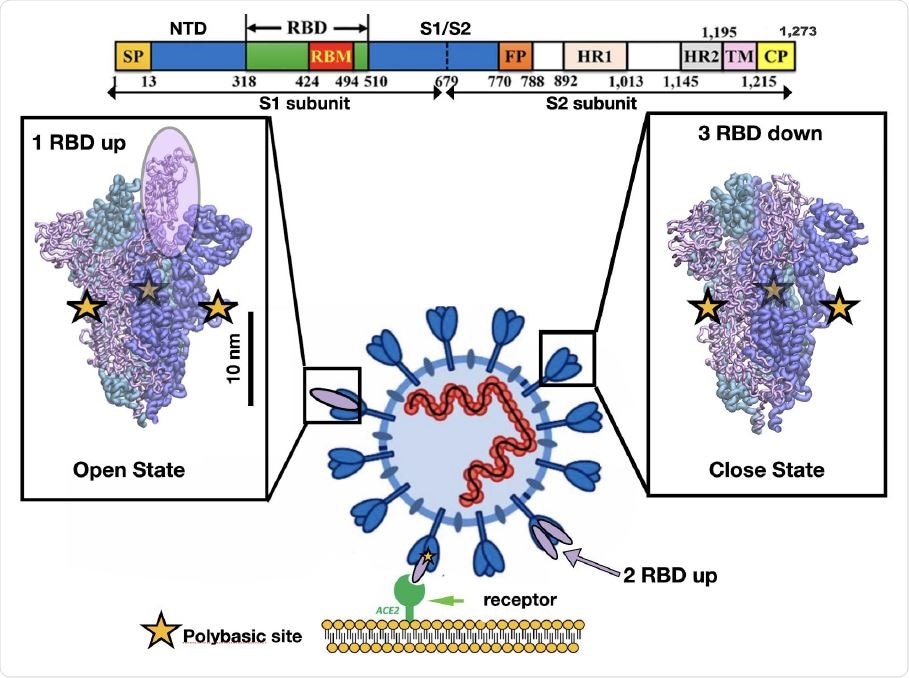
Representation of the different conformations of the RBD in the SARS-CoV-2 spike protein. Cell recognition is initiated by the RBD transition from down to up conformation and then the high affinity triggers binding between the RBD in t he up conformation and the ACE2 receptor shown in purple and green respectively. The sequence of one chain of the spike protein is shown on top as well as the residue numbers for several protein domains. The bar shows the typical length scale for the whole system. The fusion of the viral and cell membrane takes place by surface proteases that cleave each chain at the polybasic sites (yellow stars) located at the interface of S1/S2 subunits.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Different conformations of RBD
Using computational modeling, researchers now show the correlations between different conformations of the spike protein and its subunits, RBD, and the N-terminal domain (NTD).
Differential contact map analysis has revealed how the change in the number of contacts between different conformations affects the stability of a conformation.
The authors found that although the conformation with one RBD up and two down had less destabilizing residues than the two up and one down conformation, the number of stabilizing residues were similar for the two conformations. The number of destabilizing residues for the one RBD up and two RBD up conformation forming from the three down conformation is about 3% and 15% of the total number of residues. This suggests going to the one RBD up conformation results in the destabilization of fewer numbers of residues.
Upon analyzing high-frequency contacts, the researchers found 25 stabilizing residues in the down conformation, making 83 contacts which are critical for the 40 contacts made in the one RBD up case. Such transitions happen by the rearrangements of some residues at the bottom of the RBD. Identifying residues that form stabilizing contacts is important in developing drugs that target binding or disrupt viral proteins.
A previous study identified a free fatty acid-binding a hydrophobic pocket that locks the spike protein into the closed conformation. This is possible because there are few native contacts in the nearest two RBDs. The authors also found additional contacts between the up RBD chain and the NTD, helping to stabilize the conformation. In contrast, there were few interactions between the residues in the two RBD chains in the down conformation.
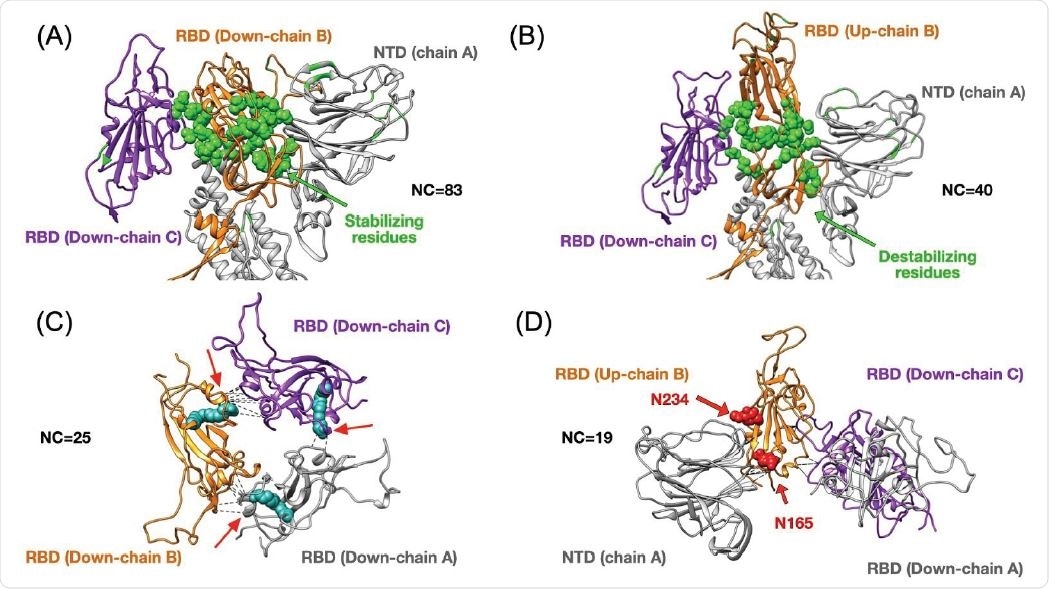
Top panels show changes in terms of stability between the closed (3down) and open (1up2down) states. Panel (A) shows the case of the RBD in chain B stabilized by neighboring protein domains such as the RBD and NTD from chain C and A respectively. The stabilizing 25 residues are highlighted in green and they establish several high-frequency native contacts (NC) equal to 83. Panel (B) shows the same set of residues from panel (A) that are destabilized in the open state forming 40 contacts. Panel (C) shows the stabilization due to 25 contacts between all RBDs in the closed state. The structure of the LA (in cyan) has been superimposed onto our results as it was shown to lock the closed state by forming contacts between two adjacent RBDs. Panel (D) depicts the RBD in up conformation that has been stabilized by 19 contacts formed between the NTD and two other RBDs. The positions of two N-glycans that assist structurally by making extensive interaction with the RBD in the up conformation have been superimposed onto our structure. We highlight the residue contacts that are responsible for stabilization by dashed black lines.
Targeting destabilization sites may inhibit viral binding
The team also determined the part electrostatic interactions play in the spike protein using the Poisson-Boltzmann method, taking into account contributions from solvation energy and Coulomb interactions.
They found that the most favorable position is when all the RBDs are in the down conformation. The conformation with one RBD up is the most favorable open position, followed by the conformation with two RBDs up. The latter conformation has a much higher energy barrier compared to the change from the closed to the one RBD up position.
The high-frequency contacts between the RBD chain and NTD chain suggest they play a role during transition from the closed to the open state. This transition occurs at an energetic cost of about 10-15 kcal/mol by breaking the bonds between these chains.
In the absence of ACE2, there is a large amount of energy required; about 30 kcal/mol to transition from the closed to the two RBD up conformations. This suggests the spike protein is likely in the one RBD up conformation before interaction with the host cell.
The information about potential target sites to destabilize the spike protein could be used to investigate potential approved drugs or herbal derivatives that can inhibit spike protein conformations that allow binding to ACE2, write the authors.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources