As the world comes to terms with the coronavirus disease 19 (COVID-19) for a long time, scientists are still exploring new approaches to reduce the mortality and morbidity of the infection in the hopes of making it a more manageable condition.
The researchers’ illuminating findings surrounding virus-host interaction present many potential routes for therapeutic development.
Translation of Viral RNA
The SARS-CoV-2 has at its core an encapsulated RNA genome; this is the virus’ genetic material that allows it to hijack human epithelium (cells lining the gut or the lungs) and use these to replicate itself. The virus also has a nucleocapsid, around which is the viral membrane containing the spike glycoprotein (or S-protein), and the viral envelope. These viral components are composed of the structural N-, M-, S- and E-proteins, respectively.
The viral genome is among the largest among RNA viruses, with a 5’-cap and a 3’-poly(A) tail. Once the virus enters the cell, the genomic RNA acts as a messenger (mRNA), which is then translated to the various enzymes and other nonstructural proteins (NSPs). These involve the 16 NSPs that form the double-membrane vesicles (DMV) vital for the further synthesis of viral RNA.
The genomic RNA (gRNA) also acts as the template for the synthesis of further strands of RNA, in 10 different canonical positive-sense forms. All these have a leader sequence at the 5’ end and common 3’ end sequences.
Of these forms, one is full-length, while the other 9 are subgenomic RNA (sgRNA). The latter are formed by discontinuous transcription, whereby the 5’ end leader sequence is fused to the downstream open reading frames that form the body of the RNA segment. These sgRNA fragments encode the structural viral proteins as well as accessory proteins.
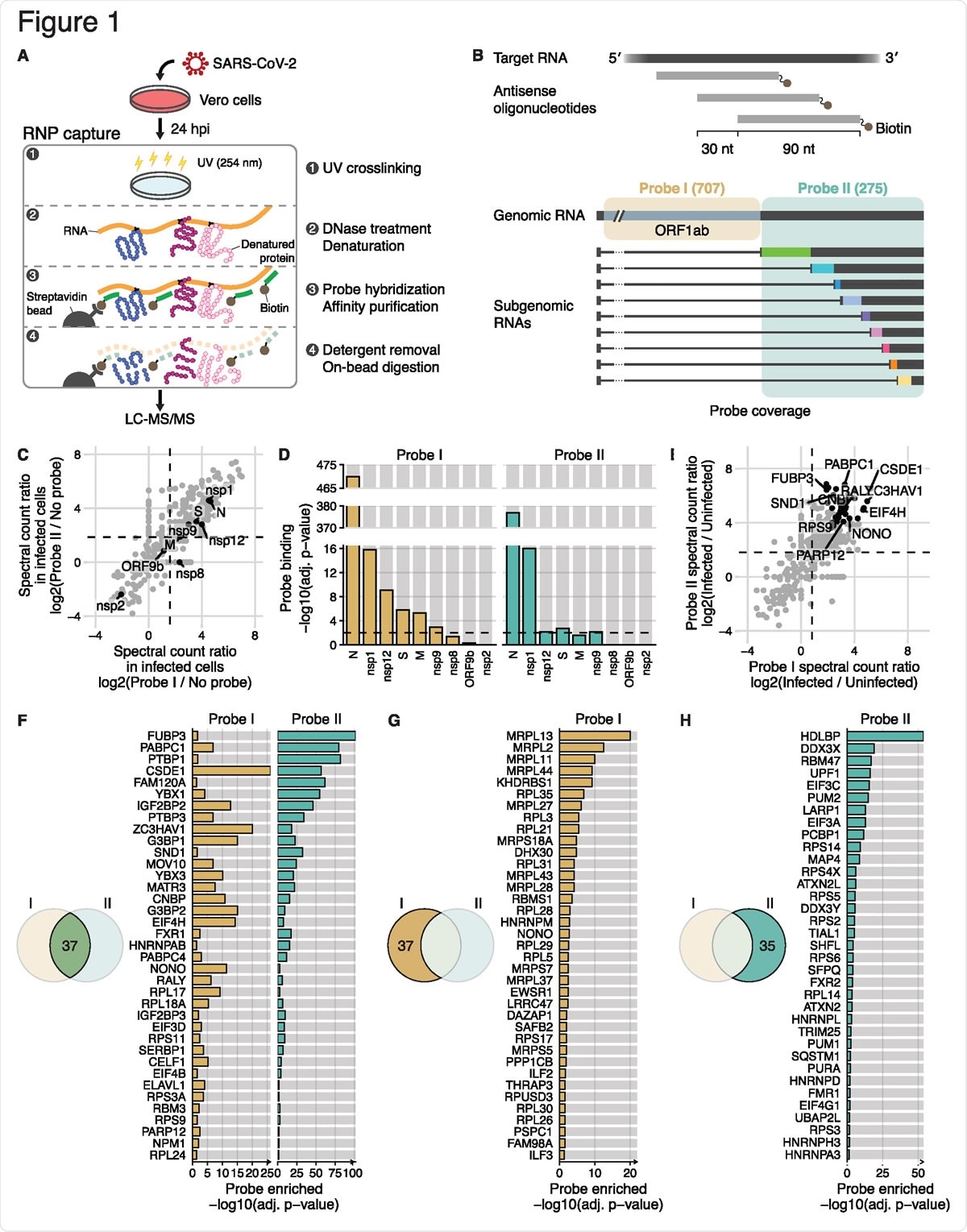
Comprehensive identification of host and viral proteins that directly interact with the SARS-CoV-2 RNAs (A) Schematic of the modified RAP-MS protocol in SARS-CoV-2-infected Vero cells. (B) Schematic of two separate pools of 90-nt antisense oligonucleotides and their SARS-CoV-2 RNA coverage. The first probe set “Probe I” consists of 707 oligonucleotides that cover the unique region of gRNA, and the second probe set “Probe II” consists of 275 oligonucleotides that cover the common region of gRNA and sgRNAs. (C) Spectral count ratio of Probe I (x-axis) and Probe II (y-axis) experiments over no-probe control in SARS-CoV-2-infected Vero cells (n = 3 technical replicates). Host proteins are marked by grey circles, and viral proteins (n = 9) are marked and labeled in black. The mean spectral count ratio of Probe I and of Probe II experiments are marked by vertical and horizontal dashed lines, respectively. (D) Statistical analysis of the quantity of viral proteins over no-probe control (i.e. probe binding). Adjusted p-values (adj. P-value) of Probe I experiments and of Probe II experiments are shown in yellow and green, respectively. The threshold for statistical significance (adj. p-value < 0.01) is indicated by horizontal dashed lines. (E) Spectral count ratio of Probe I (x-axis) and Probe II (y-axis) experiments in SARS-CoV-2- infected Vero cells compared to RNP capture experiments in uninfected cells (n = 3 technical replicates). Statistically significant host proteins (n = 37, adj. P-value < 0.05) in both Probe I and Probe II experiments are marked by black circles. Of those, representative host proteins are labeled. The mean spectral count ratio of Probe I and of Probe II experiments are marked by vertical and horizontal dashed lines, respectively. (F) Statistical analysis of host proteins enriched in both Probe I and Probe II experiment (i.e. probe enriched). Adjusted p-values (adj. P-value) of Probe I experiments and of Probe II experiments are shown in yellow and green, respectively.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Power of Regulatory RNA-Binding Proteins
All these are translated in unison with unique viral evasion and modulatory strategies that keep the process running smoothly. For instance, the gRNA is simultaneously being translated, transcribed and encapsulated. However, the host cells also respond with their own RBPs, including Toll-like receptors (TLRs), RIG-I and MDA5, which are designed to detect and clear the virus. The virus thus also encodes mechanisms to evade these host responses within its gRNA.
To accomplish this, it exploits various host regulatory proteins, such as the RNA-binding proteins (RBPs). These form specific ribonucleoprotein (RNP) complexes. The sum of these RBP-RNA interactions makes up the interactome.
Discovering these RBPs, and the way they bind to and regulate the transcribed viral RNA is important to harness the power of regulatory genes and host defenses. The current study uses an array of biochemical techniques to uncover these mechanisms at the molecular level.
These include crosslinking immunoprecipitation followed by sequencing (CLIP-seq), which studies the expressed proteins and indirectly the RNA; RNA-focused methods such as RNA antisense purification coupled with mass spectrometry (RAP-MS).
RBPs Involved in Viral RNA Regulation
The current study used a modified form of the latter to identify the full range of proteins that interact with the viral RNA, both SARS-CoV-2 and the seasonal coronavirus HCoV-OC43.
The researchers' comment, “These modifications to the RAP-MS protocol enabled robust and sensitive identification of proteins directly bound to the RNA target of interest.”
The researchers used smaller fragments of RNA, about 90 nucleotides long, as probes to identify complementary sequences on the genomic and subgenomic RNA. Two pools of probe sequences were used to identify both gRNA and sgRNA specifically.
They found 429 host and nine viral proteins, of which 199 and 220 were expressed at higher levels than in the controls. These proteins contain specific RNA-binding domains, including RNA recognition motif (RRM) and K homology (KH) domains, at higher levels compared to cellular mRNA interactomes.
In both gRNA and sgRNA, the N protein was the most highly overrepresented, with other highly expressed proteins, including nsp1, a major virulence factor. The gRNA probe detected NSP12, NSP9 (required for viral replication), and structural spike and membrane proteins more than the sgRNA probes.
When compared with uninfected cells, infected cells treated with probes I and II showed enrichment of 74 and 72 proteins, respectively. These 109 proteins are defined as the “SARS-CoV-2 RNA interactome.” The researchers also found a smaller set of core proteins interacting with the viral RNA, called the “core SARS-CoV-2 RNA interactome.” Most of these participate in regulating various viral pathways, including RNA stability and function.
Conserved Betacoronavirus RNPs
Unlike SARS-CoV-2, NSP1 in betacoronavirus lineages A and B had a different function, with only a fraction being detected in HCoV-OC43. However, 107 proteins were found in both the SARS-CoV-2 and HCoV-OC43 interactome, at different durations from the point of infection. This indicates the conservation of RBPs among coronaviruses.
Again, 14 of 38 host proteins involved in the transcriptional regulation, processing and stability of RNA, detected by both sets of probes, are conserved between betacoronavirus RNA interactomes.
RBPs May Enable SARS-CoV-2 Immune Evasion
The researchers traced the RBPs to cellular processes concerned with translation and found that they play a major regulatory role in RNA interactions, including the viral life cycle.
SARS-CoV-2 may evade immune recognition and interferon induction, the normal antiviral response, which in turn increases interferon-stimulated genes (ISGs). Therefore, the investigators also looked at the possible role of these factors in regulating viral immune evasion.
From published data they found that, following SARS-CoV-2 infection, the host factors PARP12, SHFL, CELF1, and TRIM25 are expressed at higher levels. Of these, the first and the last are core ISGs, while SHFL is upregulated in samples of lung tissue from COVID-19 patients who succumbed to the infection. PARP12 is involved in producing antiviral activity against a range of RNA viruses.
RBPs May Suppress Antiviral Pathways
Using knockdown experiments, the researchers found the host factors that mediate viral RNA binding to these regulatory proteins and other possible antiviral host factors. Knockdown of host factors, namely PARP12, TRIM25, and SHFL, that are stimulated by SARS-CoV-2 infection or treatment with beta-interferons increases viral RNA levels.
They found that the RBPs concerned with viral RNA recognition are upregulated via JAK-STAT and interferon signaling pathways and block the infection. Several of these proteins are putative anti-SARS-CoV-2 factors. Some interact with the N-protein to block antiviral responses. In this context, the interferon-suppressing activity of SARS-CoV-2 appears to help it successfully block this antiviral defense.
Again, ADP ribosyltransferases such as PARP12 are used by many viruses. The coronavirus NSP3 can remove ADP-ribose and thus reverse the effect of PARP proteins, thus promoting viral replication.
The authors say, “Based on our RNA interactome data, we hypothesize that the RNA-binding activity of PARP12 and its role in viral RNA recognition may explain the underlying molecular mechanism of its antiviral activity against SARS-CoV-2 transcripts.”
Similarly, the interferon-mediated stimulation of many RBPs may play a role in the suppression of many antiviral mechanisms. If so, regulation of the interferon pathways may be responsible for the immune escape of SARS-CoV-2. The current study also showed many novel host factors that are being identified for the first time in a viral infection, such as LARP1, a factor that may stabilize mRNAs encoding a variety of translation factors containing the 5’ TOP motif.
Implications and Future Directions
These findings may imply that SARS-CoV-2 infection can induce multiple other host factors that promote viral translation, like EIF3A, EIF3D and CSDE1. By recruiting specific factors, the virus may regulate translation initiation. Again, these interactomes are found to contain a higher level of RBPs with KH domains, whereas the cellular mRNA does not.
Other possibilities include the control exerted by RNA over the proteins interacting with it; and changes in the sgRNA/gRNA ratio since much sgRNA may be decoy RNAs to modulate the host immune response and prevent viral suppression. The broad range of interactions with antiviral factors provides a glimpse of many other potential approaches to exploring therapeutic strategies for this infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lee, S. et al. (2020). The SARS-CoV-2 RNA Interactome. bioRxiv preprint. doi: https://doi.org/10.1101/2020.11.02.364497, https://www.biorxiv.org/content/10.1101/2020.11.02.364497v1
- Peer reviewed and published scientific report.
Lee, Sungyul, Young-suk Lee, Yeon Choi, Ahyeon Son, Youngran Park, Kyung-Min Lee, Jeesoo Kim, Jong-Seo Kim, and V. Narry Kim. 2021. “The SARS-CoV-2 RNA Interactome.” Molecular Cell, April. https://doi.org/10.1016/j.molcel.2021.04.022. https://www.cell.com/molecular-cell/fulltext/S1097-2765(21)00327-0?.