Researchers in New Zealand, the United States and Australia have demonstrated the effectiveness of real-time genomic sequencing at tracking the re-emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in New Zealand, in August this year.
SARS-CoV-2 is the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic that continues to plague the globe posing a threat to public health and the economy.
Jemma Geoghegan from the University of Otago in Dunedin, New Zealand, and colleagues say real-time genomic sequencing quickly identified that the new cases belonged to a single genomic lineage and were, therefore, the result of a single introduction.
The sequencing was used to inform the lockdown measures and track and trace efforts needed to control the outbreak and enable the virus to be eliminated from the community for a second time.
However, the team also says substantial biases and gaps in global sequencing data limited the power of the genomics to successfully identify the precise origin of the August outbreak.
The researchers advise that potential sampling biases and gaps in this sequencing data should always be carefully considered when trying to identify the origin of a specific SARS-CoV-2 outbreak.
They also say that access to a broader and more heterogeneous sample of global genomic data would improve future efforts to locate the sources of new outbreaks.
A pre-print version of the paper is available on the server medRxiv*, while the article undergoes peer review.
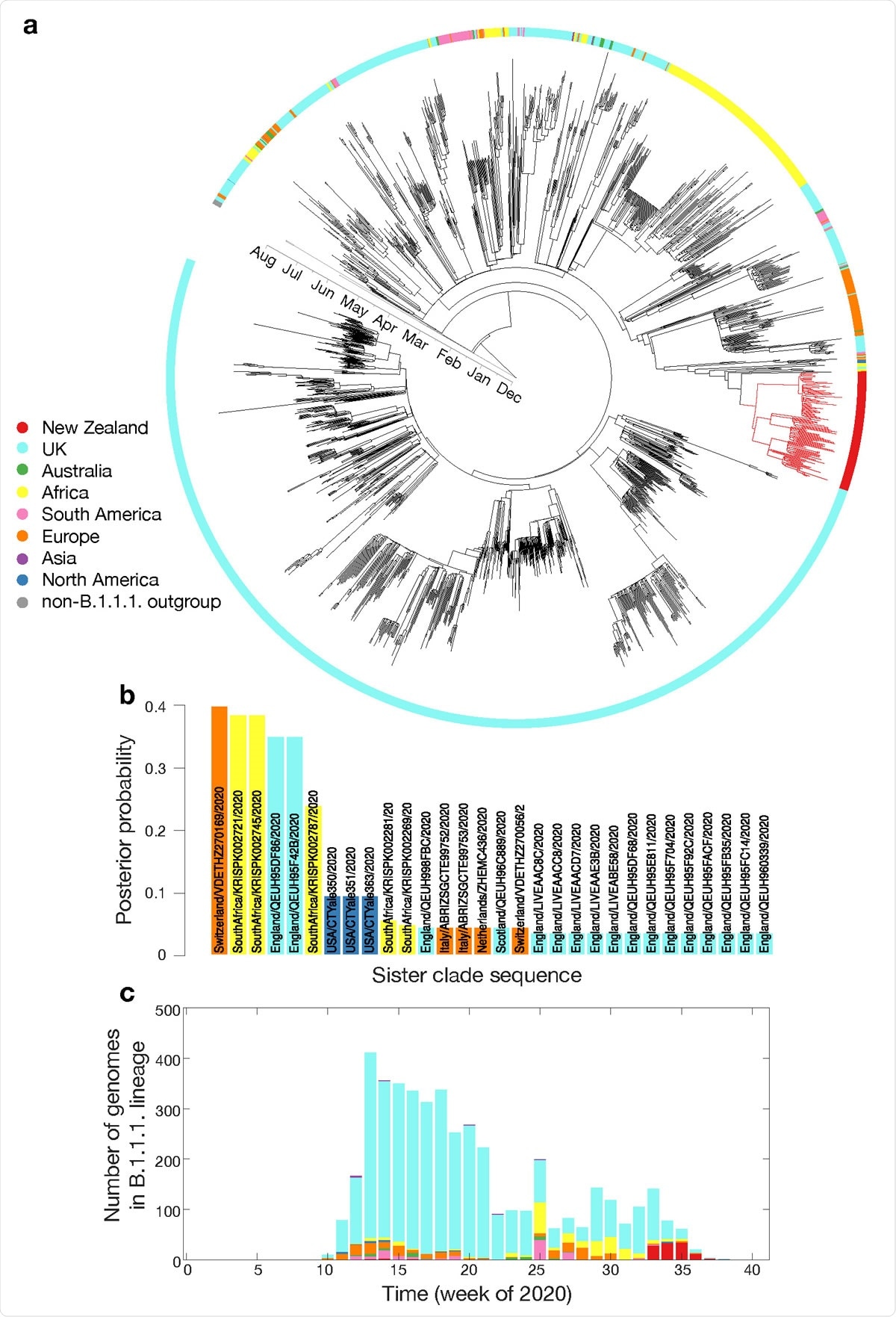
a. Maximum clade credibility phylogenetic tree of 2,000 subsampled global genomes (1,996 most recently sampled B.1.1.1. plus four non-B.1.1.1. used as an outgroup) with an outer ring coloured by sampling region; b. Posterior probability of genomes within the sister clade to New Zealand’s August outbreak, colour-coded by sampling location; c. Proportion of genomes within lineage B.1.1.1. in the global data set over time, colour-coded by sampling location.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Genomic sequencing of SARS-CoV-2 “has occurred so quickly”
Just twelve days after SARS-CoV-2 was first identified, a genome of the virus had been published, and as of September 25th this year, more than 110,000 SARS-CoV-2 genomes had been made publicly available.
“The underlying genome sequencing has occurred so quickly that, for the first time during an infectious disease outbreak, it has enabled virological and epidemiological data to be integrated in real time,” say Geoghegan and colleagues.
Real-time genomic sequencing of these data has been pivotal in informing the response to the pandemic by tracking the global transmission and evolution of SARS-CoV-2, including the identification of the number, source, and timing of introductions into different countries.
However, there has been significant between-country variation in the number and proportion of positive cases sequenced and genomes published, say the researchers.
Geoghegan and colleagues say such disparities in sequencing efforts can have important implications for data interpretation and must be met with careful consideration.
The re-emergence of SARS-CoV-2 in New Zealand
“Real-time sequencing of SARS-CoV-2 genomes has, however, had particular utility in tracking the re-emergence of the virus in New Zealand,” says the team.
Following the initial outbreak in late February, SARS-CoV-2 had effectively been eliminated in the country by June, with any positive cases limited to those linked to quarantine facilities at the border.
However, following more than one hundred days of no detectable community transmission, four new cases emerged on August 12th, none of which could be epidemiologically linked to any known case.
During this second outbreak, genomic sequencing was used to support track and trace efforts in the country for the first time.
Geoghegan and colleagues generated the genomes of 80% of the laboratory-confirmed SARS-CoV-2-positive samples from the new outbreak. They then compared these to sequenced cases from the first outbreak and to those from quarantine facilities.
However, no link was identified, and the team went on to compare the genomes from the new community outbreak to the global dataset.
What did they find?
Initial genomic sequencing was able to quickly identify that the new COVID-19 cases and subclusters were linked to the one genomic lineage B.1.1.1, therefore showing that the outbreak had resulted from a single introduction.
However, of the countries that had so far contributed SARS-CoV-2 genomic data, 40% had genomes originating from this lineage.
Phylogenetic analysis of the most recently sampled B.1.1.1. genomes found that those identified in Switzerland, South Africa, and England in August were the closest relatives of the viruses associated with the new outbreak in New Zealand.
However, genomic epidemiological analysis on the possible origins of the new outbreak was found to be inconclusive, which the team says is “likely due to missing genomic data within the quarantine border facilities as well as in the global data set.”
For example, twelve SARS-CoV-2 genomes from people returning to New Zealand from India who all arrived on the same flight spanned at least four genomic lineages, with sequence divergence of up to 34 genomic mutations.
“Such a high level of diversity in just a small sample of positive cases from India suggest that the currently available genomic data fails to encompass the true diversity that existed locally, let alone globally,” says the researchers.
Real-time genomic sequencing helped New Zealand eliminate the virus a second time
However, real-time genomic sequencing following the re-emergence of the virus helped to quickly inform the track and trace efforts and lockdown measures needed to control the outbreak, putting New Zealand on track to eliminate the virus for the second time, they add.
Nevertheless, the biased nature of global sampling clearly limited the power of genomics to identify the geographical origin of New Zealand’s August 2020 outbreak, says the team.
“We, therefore, advocate that careful consideration of the potential sampling biases and gaps in available genomic data be made whenever attempting to determine the geographic origins of a specific outbreak of SARS- CoV-2,” write Geoghegan and colleagues. “Analysis should consider all available evidence, including from genomic and epidemiological sources.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Geoghegan J, et al. The power and limitations of genomics to track COVID-19 outbreaks: a case study from New Zealand. medRxiv, 2020. doi: https://doi.org/10.1101/2020.10.28.20221853, https://www.medrxiv.org/content/10.1101/2020.10.28.20221853v1
- Peer reviewed and published scientific report.
Geoghegan, Jemma L., Jordan Douglas, Xiaoyun Ren, Matthew Storey, James Hadfield, Olin K. Silander, Nikki E. Freed, et al. 2021. “Use of Genomics to Track Coronavirus Disease Outbreaks, New Zealand.” Emerging Infectious Diseases 27 (5): 1317–22. https://doi.org/10.3201/eid2705.204579. https://wwwnc.cdc.gov/eid/article/27/5/20-4579_article.