The source of the rampant coronavirus disease (COVID-19) pandemic is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which contains a single-stranded RNA molecule as its genetic material. Accordingly, the important feature of the virus is the presence of viral mutations.
Strain-level mutations are frequently found among affected populations and bestow viral genetic fingerprints that researchers and epidemiologists may employ to trace transmission routes across broad geographic regions, which is useful in contact tracing for super-spreader events.
Likewise, less frequent de novo mutations that appear as the virus replicates are also seen in individual patients with active infections, which subsequently define sub-clonal quasispecies that is pertinent to an infected individual.
The rising availability of SARS-CoV-2 genome sequences represents a rich resource for biomedical and clinical researchers to delve deep into investigating pandemic spread. Nonetheless, traditional alignment-based analysis methods are laden with challenges when they are applied on a scale of thousands of genome sequences.
Leveraging thousands of sequenced SARS-CoV-2 genomes, a research group from Stanford University in the United States (led by Dr. Billy T. Lau) performed a viral pangenome analysis in order to pinpoint conserved genomic sequences and develop a rapid and highly scalable targeted sequencing assay.
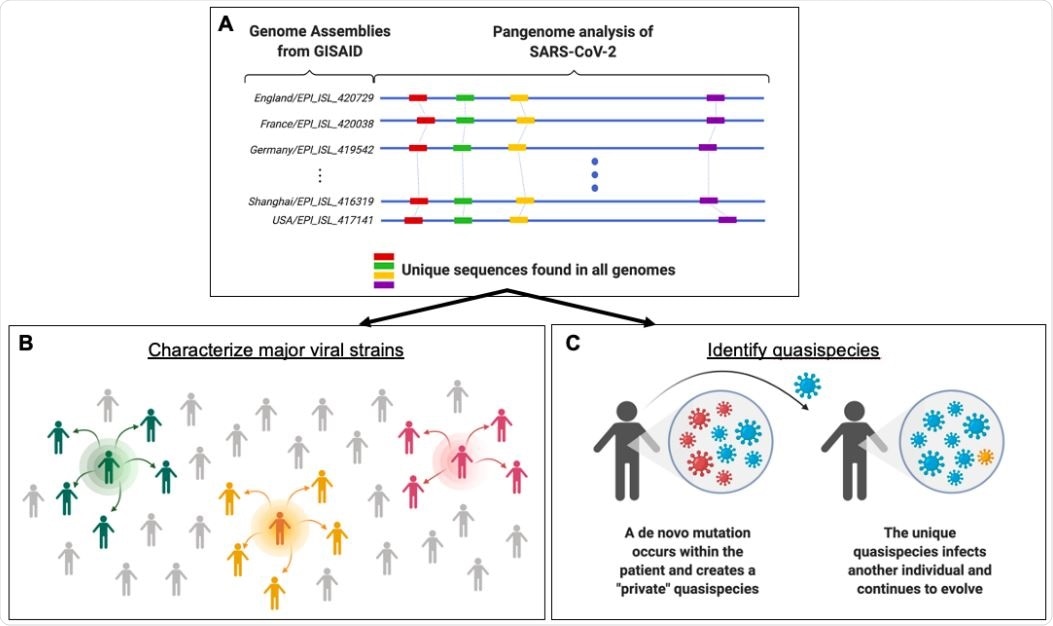
Framework for identifying population-level SARS-CoV-2 strains and lower frequency quasispecies through pangenome analysis. (A) GISAID currently has thousands of SARS-CoV-2 genomic sequences banked. By analyzing large numbers of viral genomes together (blue lines), one can pinpoint sequences that are present only once in a given genome but also occur consistently across all genomes (red, green, yellow, and purple bars). These unique and conserved sequences can be used for a number of research and clinical sequencing applications. (B) This knowledge can fuel epidemiological studies and allow scientists to characterize major SARS-CoV-2 strains. The green, orange, and pink icons represent contagious individuals who contribute to the transmission of a virus within a population. (C) The targeted sequencing enables the detection of low-frequency quasispecies that are created through de novo mutations within an individual. The mutation profile from pangenome analysis allows us to examine whether these mutations create a unique genetic profile that can be traced to and across individuals

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Identifying mutations and specific viral signatures
To identify conserved regions across thousands of SARS-CoV-2 genome assemblies, the researchers have developed a stringent computational workflow that analyzes k-mer sequences that are specific viral signatures (i.e., short tracts of sequence) – most notably when a set of genomic elements are compared.
Based on their identification of conserved regions of the SARS-CoV-2 pangenome, they have subsequently developed a sequencing assay in order to identify novel mutations. More specifically, rather than covering the entire viral genome, they have utilized highly conserved sequences as primer sites that flank highly variable genome regions.
This novel assay was made to enable very deep sequencing coverage, with thousands of reads per given base on average. As a result, researchers were able to analyze a series of contrived samples, artificially made viral admixtures, and clinical samples.
Important operational advantages of the novel assay
Consequently, this specific method enabled the researchers to swiftly survey viral pangenome characteristics (such as genome conservation and mutation signatures) from thousands of viral samples, with a relatively high amount of computational efficiency.
More specifically, evidence from their pangenome study revealed evidence of evolutionary divergence with less than 10% of all bases sharing an anchor 25-mer sequence. Naturally, additional studies will be required to determine the consequences of these changes on viral fitness and disease.
Furthermore, natural variations observed in SARS-CoV-2 have the propensity to impact the performance of primer binding and obscure mutations by imperfect primer annealing during polymerase chain reaction (PCR) procedures.
Overall, the characterization of the SARS-CoV-2 genetic variation provided significant insight into the paths of transmission and selection processes that may influence infection rates. The introduced sequencing assay shows important operational advantages compared to other molecular detection methods.
The feasibility and scalability of the approach
"Based on the results of our study, this approach has the possibility of providing a highly scalable and integrated framework for identifying viral genetic fingerprints among patients that are relatively unique," conclude study authors in this medRxiv paper.
For small genomes like SARS-CoV-2, tens of thousands of samples can be sequenced in a single sequencing run, depending on the sequencer capacity. This scalability makes the analysis of large numbers of samples feasible in comparison to other assays which necessitate maintaining samples in individual wells.
The operational scalability of next-generation sequencing also opens the door towards large-scale population screening endeavors with the potential for significant cost reduction in comparison to other methods, which is indispensable in our ongoing battle against COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lau, B.T. et al. (2020). Profiling SARS-CoV-2 mutation fingerprints that range from the viral pangenome to individual infection quasispecies. medRxiv. https://doi.org/10.1101/2020.11.02.20224816, https://www.medrxiv.org/content/10.1101/2020.11.02.20224816v1
- Peer reviewed and published scientific report.
Lau, Billy T., Dmitri Pavlichin, Anna C. Hooker, Alison Almeda, Giwon Shin, Jiamin Chen, Malaya K. Sahoo, et al. 2021. “Profiling SARS-CoV-2 Mutation Fingerprints That Range from the Viral Pangenome to Individual Infection Quasispecies.” Genome Medicine 13 (1). https://doi.org/10.1186/s13073-021-00882-2. https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-021-00882-2.