Researchers continue to expand their knowledge of coronaviruses, including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogen. Now, a new study published on the preprint server bioRxiv* in November 2020 describes a vital mechanism whereby the novel SARS-CoV-2 pathogen can restrict the human host response. In doing so, the study explores how this novel feature of SARS-CoV-2 works and how it results in a reshaping of the human host's immune response.
The researchers describe processing bodies (PBs), which are aggregations of biomolecules that occur throughout the body. These are formed via a liquid-liquid phase separation of proteins that have regions of intrinsic disorder, as well as from interactions between RNA and proteins or between different RNA molecules. They are composed of enzymes that act on messenger RNA (mRNA), producing RNA turnover by various processes.
The RNA in PBs is delivered there by RNA-binding proteins (RBPs). PBs are actually granules of ribonucleoprotein (RNP) and undergo constant fluctuations in size and number as they respond to various stimuli. For instance, anything that activates the p38/MK2 MAPK pathway causes disassembly of the PBs, as do viral infections.
The RNA within PBs makes up about a third of all coding but imperfectly translated RNA, as well as noncoding RNA. Transcribed mRNA within the PBs consists mostly of regulatory mRNAs in groups related by function.
One group includes growth factors, inflammatory cytokines and vascular growth factors. This group carries destabilizing AU-rich elements (AREs) at the 3'-untranslated region (UTR). This group was subject to a higher rate of degradation and suppression when PBs were visible, while constitutive expression of ARE-mRNA was restored with the loss of PBs. This is important in view of their physiological function.
Thus, the ARE-mRNA mechanism allows cells to respond to stimuli like viral infections that result in the PBs being lost, with the secretion of ARE-containing inflammatory cytokines at a high level, including IL-6, IL-8, IL-1β, and TNF.
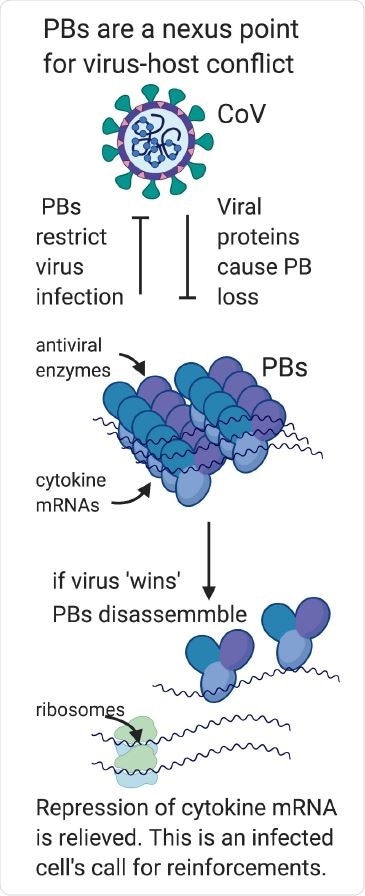
Processing bodies are a nexus point for virus-host conflict. In part one of our model, we hypothesize that PBs are direct-acting antiviral granules that can restrict virus infection when present as visible condensates; for this reason, they are targeted for disassembly by most viruses. In part two of our model, we propose that viral PB disassembly is perceived by the cell as a danger signal and relieves suppressed cytokine transcripts to produce proinflammatory cytokines that recruit and activate immune cells. In this way, PB disassembly may also contribute to the pathogenic cytokine responses that underly many viral illnesses including COVID.
Effect of viral proteins on PBs
Disassembly of PBs may be through the direct effect of a viral protein on a PB component, which then moves towards viral replication and transcription pathways within the cell, or is broken down by viral proteases. However, indirect disassembly can occur in response to viruses via p38/MK2 signals.
The assembly of PBs by viral gene products is much less common. This may indicate that PBs are directly antiviral and that the tearing down of these particles is virus replication-friendly. This is in contrast to other RNPs like stress granules, which have a more indirect antiviral activity, mediated by the enzymatic activity of the PBs.
Another indirect antiviral effect of PBs is by their stockpiling of proteins involved in innate immune signaling pathways. More studies will be needed to identify the importance of the organization of proteins within the PBs or the higher-order condensation of multiple proteins within a PB in modulating antiviral PB effects.
SARS-CoV-2 hijacks antiviral responses
While all coronaviruses are known for their ability to take over host interferon and antiviral responses, SARS-CoV-2 is exceptionally skillful at doing this. The interferon response's misdirection is turning out to be one of the most important processes that determine the clinical outcome of COVID-19.
More evidence is piling up to show how this virus defeats the host's antiviral measures, such as four nonstructural proteins (NSPs) that have been recently reported to act upon specific cellular RNAs to limit IFN-β secretion and thus favor viral spread steeply.
CoVs disassemble PBs
The current study is the first to show that human coronaviruses can disassemble PBs specifically. The viral RNA can translate to several independent proteins that can cause PB loss. The formation of PBs limits later infection with the seasonal coronavirus, OC43, in many ways, by hindering nucleocapsid protein production and preventing the production of infectious progeny. Thus, PBs are a significant component of antiviral cellular responses in coronavirus infection.
SARS-CoV-2 genes that cause PB Loss
The researchers screened SARS-CoV-2 genes for those which mediate the loss of PBs and found six genes that might cause PB loss. These included the nucleocapsid (N) protein, NSP1, which shuts off host immunity, NSP6 and NSP11, and the accessory proteins ORF7b and ORF3b.
They found that two of these also markedly enhanced PB disassembly, namely, NSP1 and ORF7b. They then looked at the expression of these PB-disassembling genes in endothelial cells and found N, (an RBP that induces the assembly of viral particles) and NSP14 were associated with PB loss. With its multiple and indiscriminate RNA-binding sites, it could well pull RNA from the cytoplasm and thus prevent phase separation of PBs by physically preventing RNA-protein interaction.
The researchers also found NSP14, an enzyme that has two different functionalities. Though NSP14 caused PB loss in endothelial cells, it failed to do so in the laboratory cell line (called HeLa cells), perhaps because of competing PB-stabilizing factors in the latter. Another explanation may be that all endothelial cells studied here were selected for NSP14 positivity, while HeLa cells were not selected. Both cell lines showed some mysterious dots, which were not PBs, and these remain to be identified. However, the researchers suggest that this protein may be a stabilizer or translation promoting factor for cytokine mRNA found within PBs and containing AREs – and could thus enhance inflammatory cytokines such as TNF and IFN-β.
The NSP1 protein is the earliest viral protein to be expressed after viral entry and is also known to restrict interferon expression as well as interferon-stimulated genes (ISGs). To achieve this, it binds to the ribosomal 18S subunit to block the mRNA entry channel within the 40S subunit.
ORF7b and ORF3b are known to oppose interferon responses to viral infection or to activate the kinase p38, which elicits PB disassembly. The scientists are working on ways to examine further the roles played by NSP6 and NSP11.
When endothelial cells were infected with human coronaviruses, PB loss occurred, with much lower levels of PBs at 12 and 24 hours post-infection. This was accompanied by a 20-fold rise in the levels of IL-6 and IL-8 mRNA.
PBs show strong antiviral effects
The presence of PBs was also found to restrict infection by coronaviruses. Cells to which PBs were delivered prior to viral infection with OC43 showing limited viral replication. This, say the researchers, provides "a compelling reason why SARS-CoV-2 would coordinate an attack on cellular PBs using multiple viral proteins."
The mechanism of restriction by the PBs remains to be clarified. Moreover, the researchers wish to determine if the technique used to increase PB levels within the treated cells itself produced antiviral effects or caused the formation of larger PBs. Moreover, it is a moot question whether the proteins localized to the PB have antiviral activity irrespective of whether they are within a formed PB or not.
The critical role of PBs in immune modulation in COVID-19
The authors say, "We consider that the antiviral restriction promoted by PB localized enzymes requires the granule formation for optimal function. By that definition, factors that promote PB condensation may also be antiviral."
In that case, the formation of PBs could be targeted by the virus, since only when they are present in visible condensate form are they capable of restricting viral infections.
Finally, the breakdown of PBs prevents the normal suppression of cytokine ARE-containing mRNAs. This means that viral infection stimulates PB loss signals to the immune system as well, indicating danger. In this case, the loss of PBs could be crucial for the cellular response of the innate immune system and an automatic part of the immune activation-signaling pathway set off by viral infection.
When this signal is combined with the inability to mount an interferon response, as seen with SARS-CoV-2 infection, this could cause dangerously high and dysregulated cytokine secretion. Thus, the study concludes, PBs form a meeting point for both direct antiviral responses and proinflammatory cytokine induction, making them "a central player in the antiviral response that coordinates the immune reshaping that occurs after CoV infection."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Robinson, C.-A. et al. (2020). Human Coronaviruses Disassemble Processing Bodies. bioRxiv preprint. doi: https://doi.org/10.1101/2020.11.08.372995, https://www.biorxiv.org/content/10.1101/2020.11.08.372995v1
- Peer reviewed and published scientific report.
Kleer, M., Mulloy, R. P., Robinson, C.-A., Evseev, D., Bui-Marinos, M. P., Castle, E. L., Banerjee, A., Mubareka, S., Mossman, K., & Corcoran, J. A. (2022). Human coronaviruses disassemble processing bodies. PLOS Pathogens, 18(8), e1010724. https://doi.org/10.1371/journal.ppat.1010724. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010724.