The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the COVID-19 pandemic, spreads mainly by airborne transmission, either in respiratory droplets or aerosols. Fecal transmission is one more possible route by which the virus spreads.
Another spreading method is when people touch virus-containing droplets that fall on various surfaces like tables, doorknobs, and counters. SARS-CoV-2 has been detected on a variety of surfaces, including plastic, cardboard, steel, and metals.
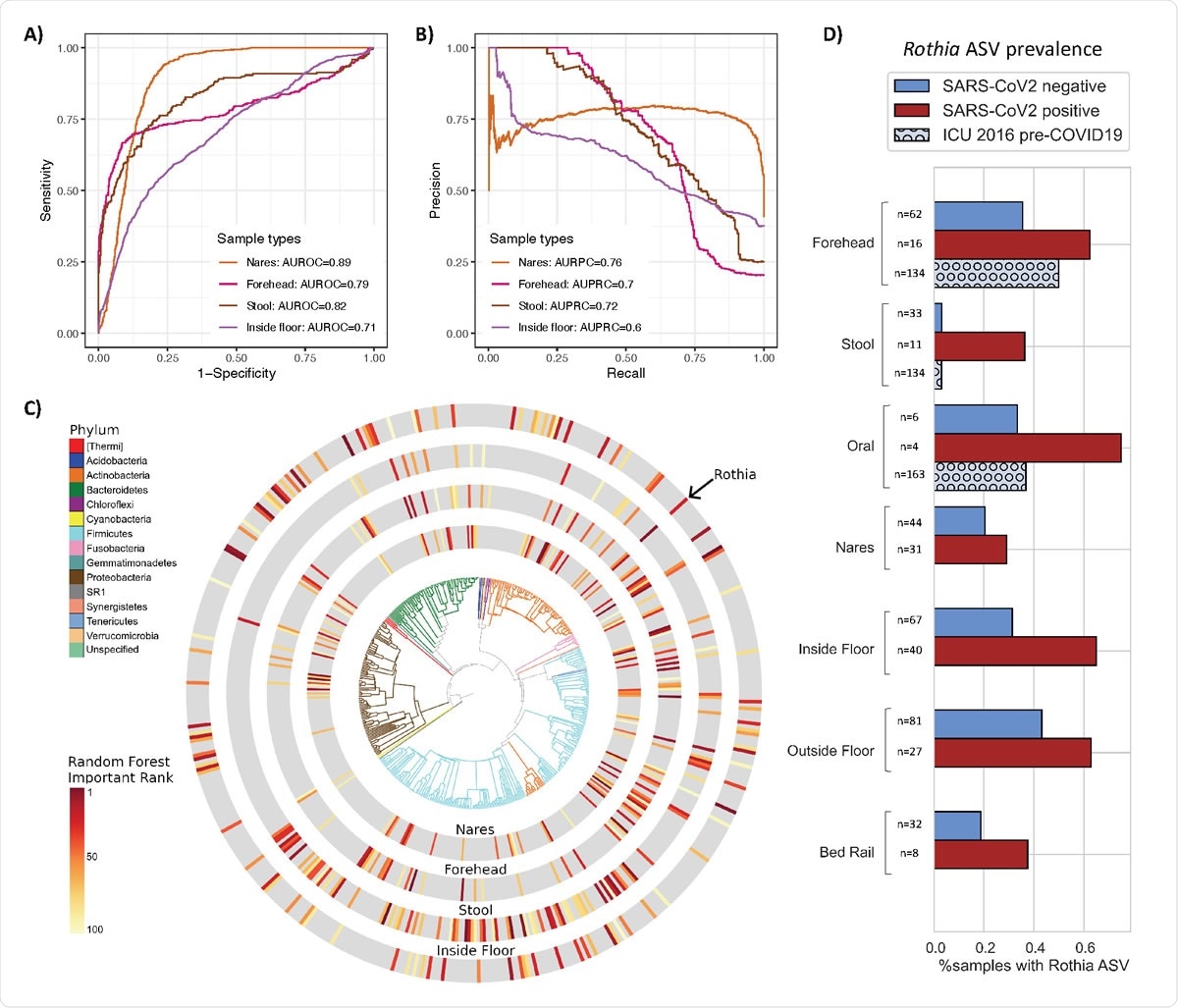
Bacterial composition is predictive of SARS-CoV-2 status in nares, forehead, stool and inside floor samples. The prediction performance of Random Forest classifiers on SARS-CoV-2 status for each sample type was assessed using AUROC (A) and AUPRC (B) for nares (n=76), forehead (n=79), stool (n=44), and inside floor (n=107), in a 100-fold cross-validation approach (see methods). (C) EMPress plot of the 100 features most predictive of SARS-CoV-2 status in nares, forehead, stool and inside floor samples, where a single ASV with 100% alignment to Rothia dentocariosa was identified across all sample types. Top 100 random forest importance ranks and GreenGenes taxonomy from nares, forehead, stool, and inside floor samples are available in S2. (D) Proportion of samples containing the highly predictive Rothia dentocariosa ASV in SARS-CoV-2 positive and negative samples from the current study, and from (30) (ICU 2016 pre263 COVID19)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The risk of contracting the SARS-CoV-2 virus is greater indoors than outdoors, especially in poorly ventilated areas. An indoor microbiome can be unique depending on the environment, mostly dominated by human-shed microbes.
The microbes generally are bacteria, but indoor areas can also have viruses when sick people are present. The interaction of virus and bacteria has been known and studied, for example, in animals, the interaction between gut bacteria and intestinal viruses. Gut bacteria can affect gastrointestinal virus infectivity by improving the thermostability and environmental stability of viruses. Virus-bacteria interactions have also been observed in the upper respiratory tract. Hence, it is possible that such interactions are present even in indoor spaces.
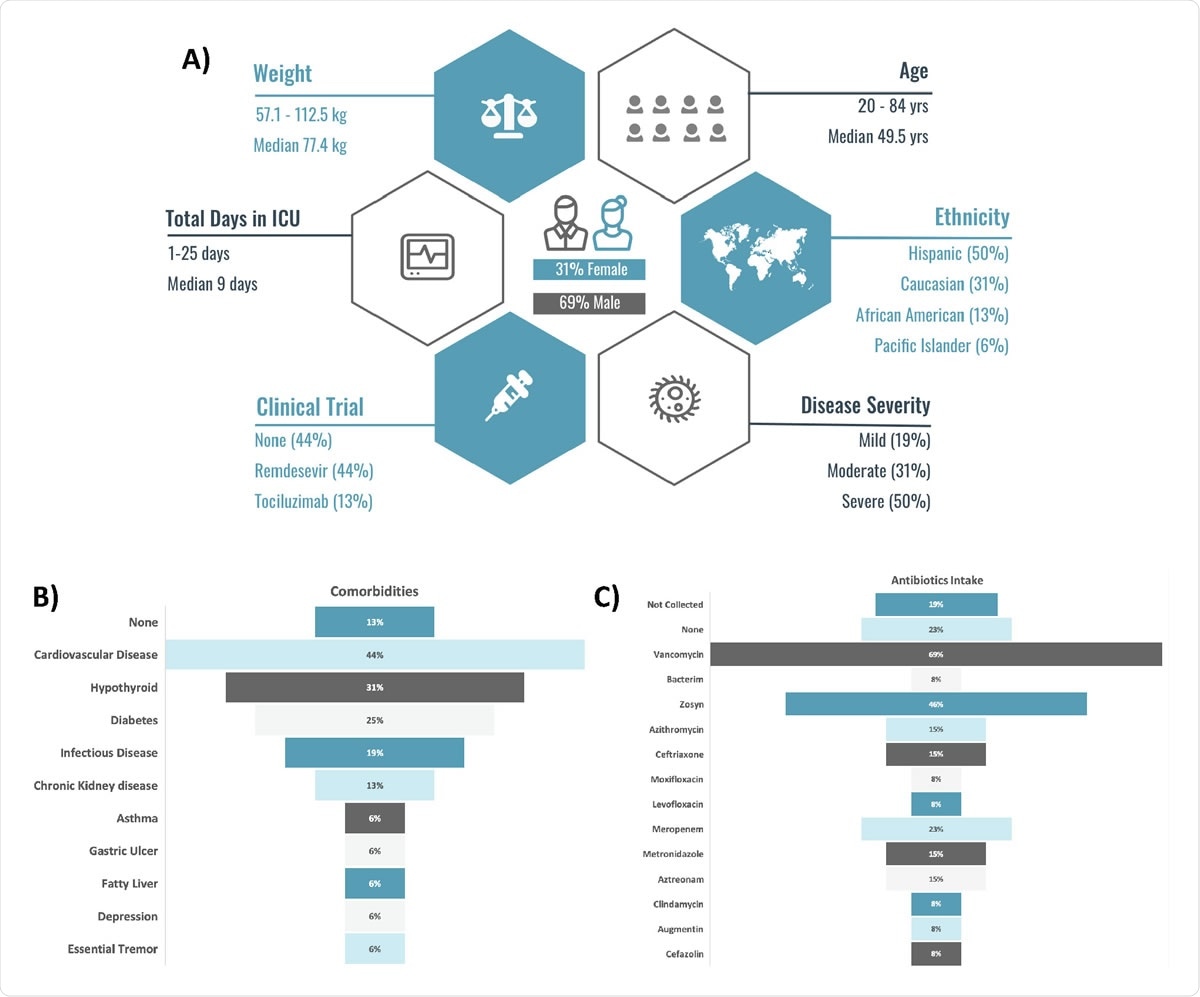
Patient (n=16) demographics (A), antibiotics intake (B), comorbidities (C).
Microbiome in SARS-CoV-2 patient rooms
Researchers from the University of California, San Diego, IBM, T. J. Watson Research Center and IBM-Almaden explored if the microbes present in an indoor environment can affect SARS-CoV-2 infection. They reported their results in a paper published on the medRxiv* preprint server.
The authors obtained swab samples from patients and healthcare workers at the UCSD Medical Center. They swabbed the skin, respiratory tract, stool, and various surfaces the patients and healthcare workers came in contact with to collect 972 samples from people and 734 samples of hospital surfaces.
Of the surface samples, about 13% were positive for SARS-CoV-2. Floors near a patient’s bed had the most virus. Some virus was also found in patients who had tested negative for the virus and in rooms cleaned after being occupied by a positive patient.
Using the 16S rRNA test, they identified the different microbes in the samples. They found different clusters of microbes on the floor, stool, and nostril/forehead samples. Surfaces frequently touched by healthcare workers had microbiomes similar to their microbiomes, and those touched by patients more had microbiomes similar to the patients.
The average species diversity was much higher in the surface samples, especially the floor, than in samples from people, with the diversity being more in SARS-CoV-2 positive samples. Analyzing the microbes present in positive samples, the authors found Clostridiales to be a group present in high proportion the stool samples, similar to that found in wastewater studies. Actinomyces, Anaerococcus, Dialister, Gemella, and Schaalia were seen in the forehead and nostril samples.
They found Rothia dentocariosa to be common on the forehead, nostril, stool, and floor samples and was more prevalent in SARS-CoV-2 positive samples. Rothia is generally not present in hospital samples, found the authors, after comparing a previous hospital microbiome study, and was specific to SARS-CoV-2.
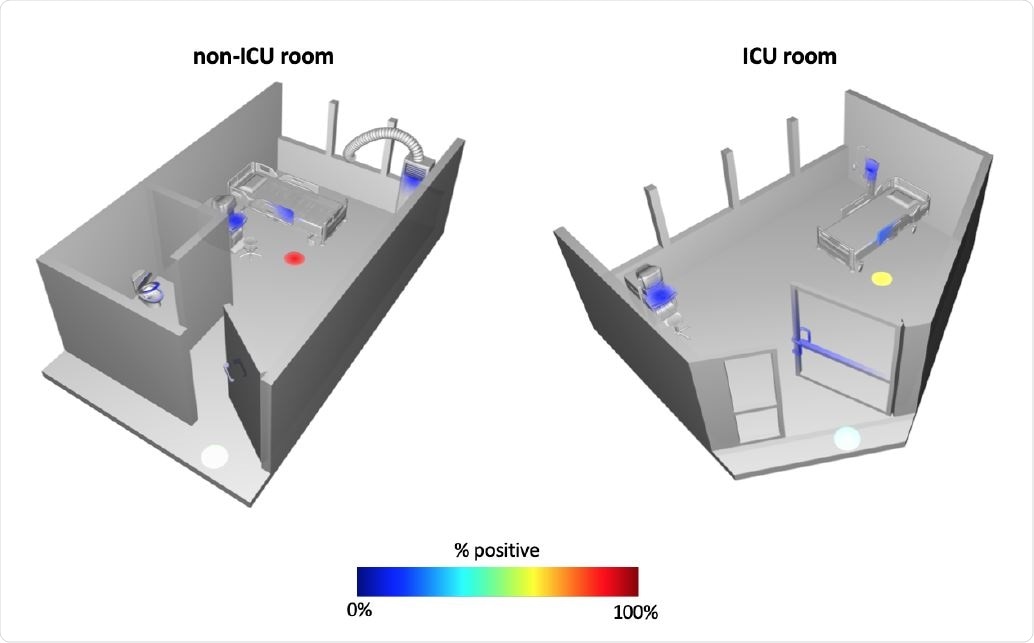
Ili’ spatial mapping of standard hospital (non-ICU) room and intensive care unit (ICU) room. Heatmap depicts the percent of samples collected at each site that were positive for SARS CoV-2.
Association between SARS-CoV-2 and indoor bacteria
Although the SARS-CoV-2 virus was detected in the indoor environment in patient rooms, it is unknown if the viruses detected were viable. Because of the high Ct values of most of the samples and lack of infection in the healthcare workers caring for the patients, the authors conclude that positive surfaces were unlikely sources of viral infection in the hospital setting studied when appropriate personal protection was used. However, effective cleaning practices should be maintained to routinely clean surfaces to lower the chance of transmission.
The presence of Rothia has been seen in previous studies on SARS-CoV-2, suggesting a strong association with the virus, although the mechanism is not clear. Rothia species have been seen in the human oral and gastrointestinal microbiome and suggests increased oral-fecal transmission, a characteristic of COVID-19. Rothia has also been seen to be higher in COVID-19 patients with cardiovascular disease, but it has also been observed in cardiovascular patients without COVID-19. More studies are needed to understand this association and if it can be used in methods to reduce SARS-CoV-2 transmission.
Among all the samples, forehead, nostrils, stool, the bacterium Rothia dentocariosa was highly predictive of and associated with SARS-CoV-2 infection. The association could be a result of direct interaction of the bacteria with the virus or indirectly through effects on the host, suggesting there is perhaps a role of bacteria-virus synergy in COVID-19 disease transmission.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Marotz, C. et al. (2020) Microbial context predicts SARS-CoV-2 prevalence in patients and the hospital built environment.medRxiv. https://doi.org/10.1101/2020.11.19.20234229, medrxiv.org/content/10.1101/2020.11.19.20234229v1
- Peer reviewed and published scientific report.
Marotz, Clarisse, Pedro Belda-Ferre, Farhana Ali, Promi Das, Shi Huang, Kalen Cantrell, Lingjing Jiang, et al. 2021. “SARS-CoV-2 Detection Status Associates with Bacterial Community Composition in Patients and the Hospital Environment.” Microbiome 9 (1). https://doi.org/10.1186/s40168-021-01083-0. https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-021-01083-0.