Wastewater surveillance has been reported to be a feasible and efficient way to monitor the prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the current COVID-19 pandemic. A recent study draws attention to the need to freeze wastewater samples if the analysis is carried out later rather than immediately.
Wastewater analysis is not only a useful monitoring tool for current prevalence but helps to predict future outbreaks early by a warning increase in wastewater counts of the virus. Such analysis has also led to the tracing of the virus to its first appearance in many of the world's cities. In fact, many reports have already traced SARS-CoV-2 in many locations worldwide before the first reports from Wuhan, China.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This is leading to intensive attempts to identify the location where the virus made its first appearance. At present, stored samples can be analyzed as wastewater testing resources are becoming more readily available. However, the pandemic's ongoing second wave may necessitate further restrictions, which could again delay sample analysis.
The impact of storage conditions on the results of testing is an important parameter that helps determine the tests' reliability. As of now, direct information on the persistence of the virus in stored wastewater samples is available only for temperatures between 4°C and 37°C. Studies on other coronaviruses have added related information.
The current study, published in the preprint server medRxiv* in November 2020, presents a solution to the frequent delay in the processing of wastewater aliquots during COVID-19. The reasons for such delay are manifold, ranging from scarcity of staff, closure of laboratories, lack of reagents and labware, and a poor understanding of the best techniques. For this reason, it has often been necessary to store the samples for later analysis.
This study is the first Finnish study designed to detect SARS-CoV-2 in municipal wastewater. Its objective was to uncover the stability of SARS-CoV-2 RNA in colder temperatures. This would help assess the accuracy of the results in many wastewater studies already published.
The researchers used an E-Sarbeco RT qPCR assay to measure the titer of the SARS envelope gene, and the N2 assay for the SARS-CoV-2 nucleocapsid gene over time. The wastewater samples examined had been stored at 4ᴼC, -20ᴼC, and -75ᴼC.
The wastewater treatment plant sampled was the Viikinmaki plant that serves 860,000 people in eight municipalities. Influent samples were taken prior to any treatment, in a 20-liter container, being taken from the composite influent collected over 24 hours. The 20-liter sample was stored at 6°C, and the other samples of 5 and 2 liters at two different time points, to reach the laboratory within 26 hours. The small samples were then divided into 30 mL aliquots and kept at 4°C, -20°C, and -75°C until they were tested.
In the first 28 days, the RNA copy number went unmeasured due to a lack of the methodology required for analysis. The quantity was taken to be similar to the measurement taken over the first month of storage in the cold, in another sample collected after 5 weeks.
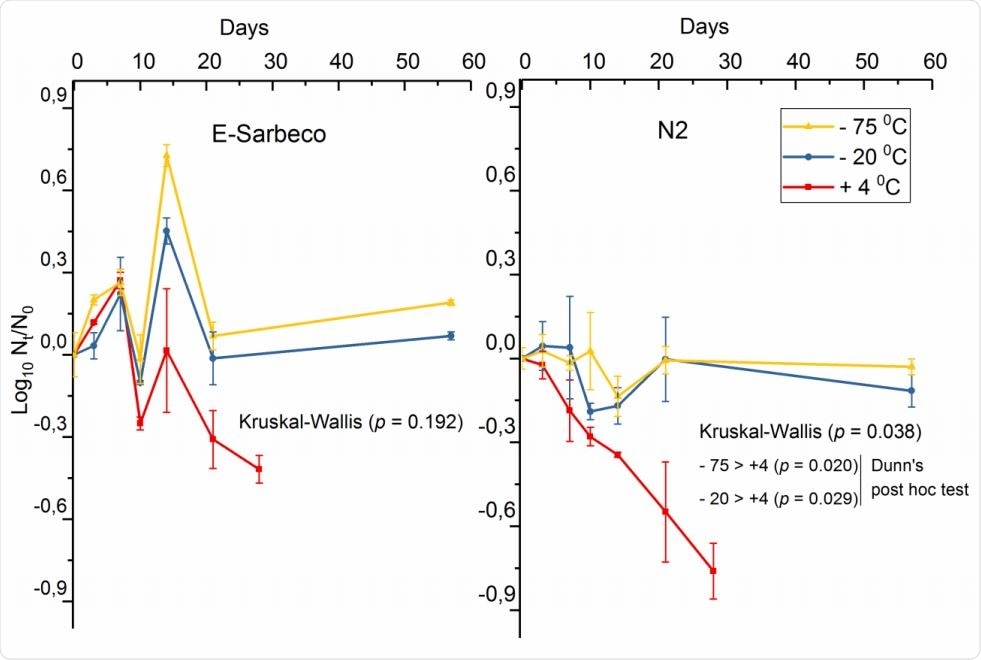
The decay curve of SARS-CoV-2 spike (log10 copies 100 ml-1) in wastewater influent at 4°C, -20°C, and - 75°C, enumerated with E-Sarbeco and N2 RT-qPCR assays.
Between 29 and 84 days, the researchers found that under all these conditions, the viral RNA remained almost stable. This was in contrast to the 1-log reduction in norovirus GII RNA during this period. This shows that non-enveloped viruses do not always persist for longer durations in the cold, as was previously thought.
Using the E-Sarbeco assay, the researchers observed similar RNA amounts irrespective of storage temperature, with a trend towards the detection of higher quantities of viral RNA at freezing temperatures. At 4°C, there was a linear decay in RNA copy number for the gene targets detected by both assays over 21 days.
The N2 assay showed a significantly greater decay at 4°C. This suggests that nucleocapsid and envelope genes have differential rates of decay. However, further study is required to understand the stability of different marker genes required for SARS-CoV-2 surveillance in wastewater.
At freezing temperatures, -20°C and -75°C, the RNA count in the influent wastewater remained intact over 58 days. The results were validated by adding a nasopharyngeal swab sample from a COVID-19 patient, in dilution, to the wastewater influent. The SARS-CoV-2-spiked sample contained stable RNA content over 28 and 58 days, the first in refrigerated and the second in freezing conditions.
While the prime determinant of viral inactivation is the storage temperature, other factors that may have a significant impact include the presence and amount of organic matter and whether other microbes are present.
There is also a need to detect low titers of viral RNA in the wastewater, which requires high-quality and efficient sampling, preserving, and processing procedures. This is particularly essential in view of the fact that previously tested viruses in wastewater have been non-enveloped viruses, while SARS-CoV-2 is enveloped.
Again, the fraction of the sample to which the virus is associated should be definitively identified by more research, whether in the pellets in the wastewater ultrafiltrate before centrifugation or particulate matter-free water fraction. The current study found slightly higher quantities of viral RNA in the former, perhaps because of the viral envelope. Some recent research, indeed, suggests the use of wastewater sludge for viral detection.
This study is important in providing data that can develop reliable protocols for wastewater testing in COVID-19 epidemiology.
The researchers conclude, "The SARS-CoV-2 RNA seemed surprisingly stable in these cold storage temperatures over 29, 64, and 84 days," at 4°C, -20°C, and -75°C. Thus, if immediate analysis is impossible, these temperatures should be preferred for storage.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hokojarvi, A.-M. et al. (2020). The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. doi: https://doi.org/10.1101/2020.11.18.20234039. https://www.medrxiv.org/content/10.1101/2020.11.18.20234039v1
- Peer reviewed and published scientific report.
Hokajärvi, Anna-Maria, Annastiina Rytkönen, Ananda Tiwari, Ari Kauppinen, Sami Oikarinen, Kirsi-Maarit Lehto, Aino Kankaanpää, et al. 2021. “The Detection and Stability of the SARS-CoV-2 RNA Biomarkers in Wastewater Influent in Helsinki, Finland.” Science of the Total Environment 770 (May): 145274. https://doi.org/10.1016/j.scitotenv.2021.145274. https://www.sciencedirect.com/science/article/pii/S0048969721003405.