Researchers in the UK conducting a genomic analysis of severe acute respiratory virus syndrome coronavirus 2 (SARS-CoV-2) and SARS-like coronaviruses in the bat and pangolin have identified strong host-associated divergences that could provide clues about the ancestry and interspecies transmission of SARS-CoV-2.
SARS-CoV-2 is the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic that continues to pose a significant ongoing threat to human life and the worldwide economy.
The study also identified a number of high-impact variants in several bat and pangolin coronaviruses that could be of functional relevance in the design of therapies and vaccines for SARS-CoV-2.
The team – from the University of Edinburgh and Aberystwyth University in Wales – says the evolutionary origins of the virus remain elusive and understanding its complex mutational signatures could guide vaccine design and development.
“Through employing a number of genomic analysis methodologies, this study has aimed to bring understanding of the diversity across SARS-CoV-2 and SARS-CoV-2-like coronaviruses by comparing a wide selection of available genomes from the starting point of the pandemic,” write Barbara Shih (University of Edinburgh) and colleagues.
A pre-print version of the paper is available on the bioRxiv* server, while the article undergoes peer review.
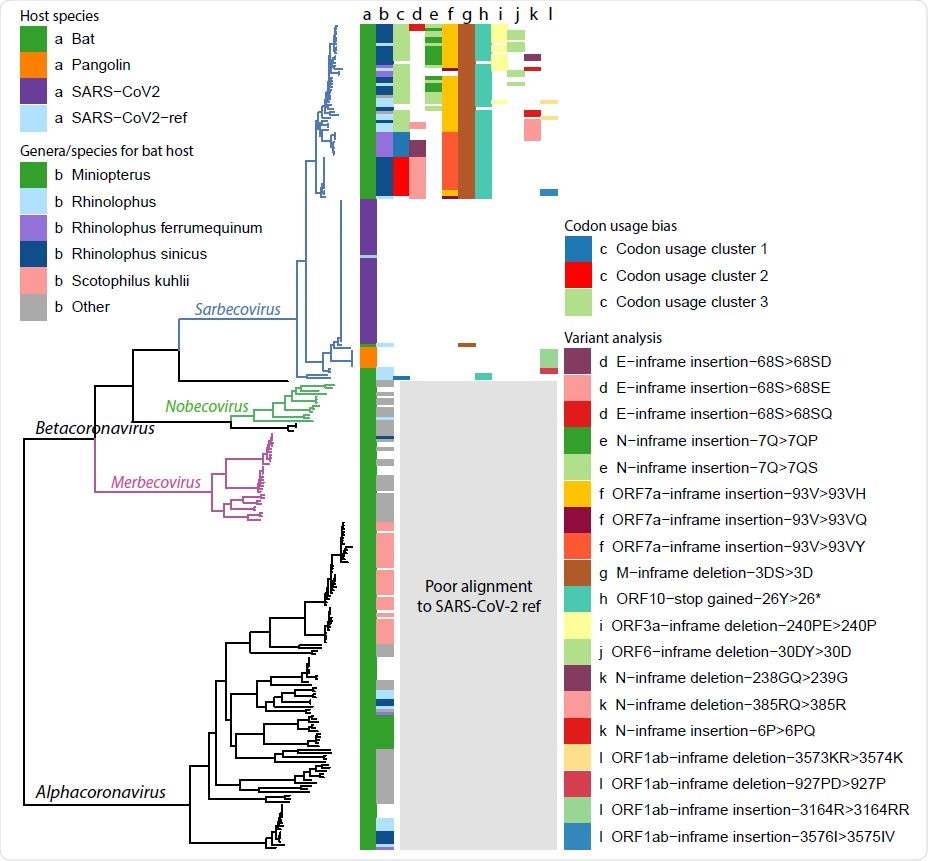
Ladderised phylogenetic tree of bat-CoV, pangolin-CoV and SARS-CoV-2 (Wuhan dataset and reference) genomes. Metadata are indicated on the top left corner, including a) dataset name and b) the bat genera and species if the genome is of bat host. Clades for Betacoronavirus subgenera, Sarbecovirus, Nobecovirus and Merbecovirus, are indicated on the graph, showing that our codon usage bias and variant analysis results are restricted to the Sarbecovirus due to poor alignment between SARS-CoV-2 ref and genomes outside this subgenera. There also appears to be some degree of genera and species separation for bat hosts. The majority of the Sarbecovirus affect the bat genus Rhinolophus (column b, light blue, dark blue and purple), whereas a much smaller proportion of the Alphacoronavirus are found in bats of this genus. Some clades overlap with specific bat species, including Rhinolophus ferrumequinum, Rhinolophus sinicus and Scotophilus kuhlii. The results from the analysis made in later parts of this study are also highlighted, including c) codon usage bias clusters, d-f) high impact variants with multiple variants are found in the same amino acid position, g-j) other high impact variants with a single amino acid change found in > 10 genomes, k-l) other high impact variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Researchers have been trying to understand the ancestry and transmission of SARS-CoV-2
Since SARS-CoV-2 first emerged in Wuhan, China, late last year (2019), significant efforts have been made to understand its transmission and how it might be contained and treated.
Coronaviruses (CoVs) are a family of large single-stranded, enveloped RNA viruses that can be divided into four subfamilies: the alphaCoVs, betaCoVs, gammaCoVs, and deltaCoVs. Like SARS-CoV-1 and Middle East respiratory syndrome (MERS) CoV, SARS-CoV-2 belongs to the betaCoV subfamily.
The CoVs exhibit at least six open reading frames (ORFs) and four structural proteins: membrane (M), nucleocapsid (N), envelope (E), and spike (S) – the latter being the main surface structure the viruses use to enter host cells.
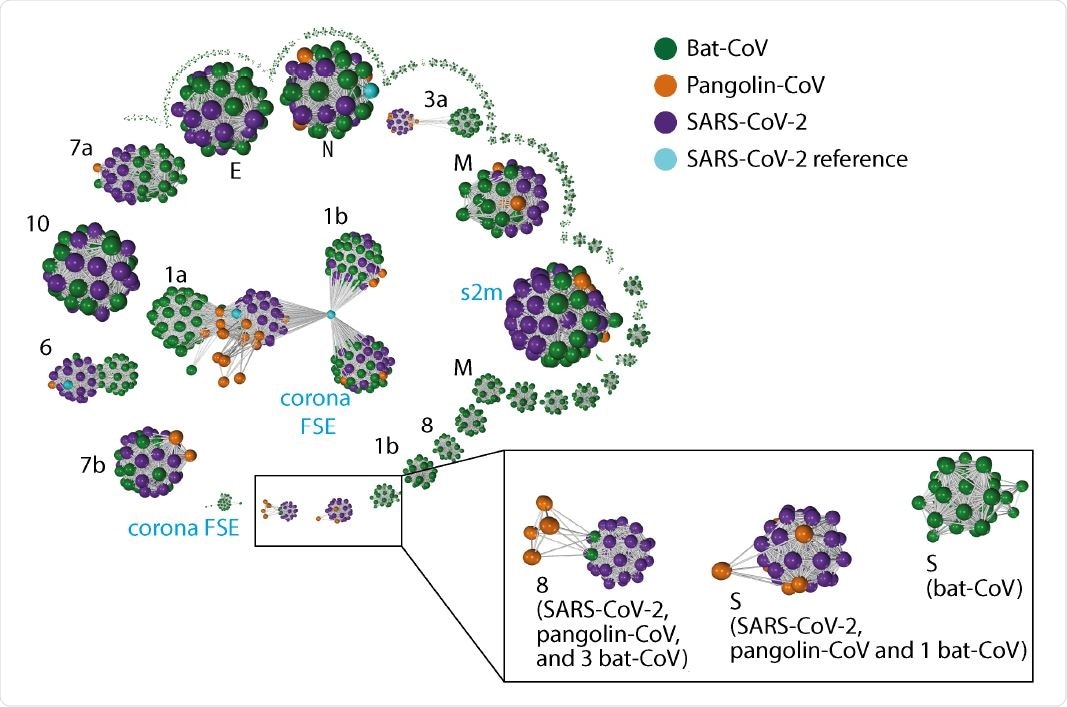
Gene-gene similarity network analysis. Each node represents a gene defined by PROKKA or a DNA segment similar to genes from the SARS-CoV-2 reference genome. The nodes were compared against each other using BLAST, and nodes with high similarity (BLAST score g 60 and a query coverage g 80%) were connected with an edge. The network graph is labeled with host species. The black font in the graph indicates the corresponding SARS-CoV-2 gene names (“ORF” omitted) for the larger clusters, whereas blue font indicate additional non-coding sequences de�ned by PROKKA. Instead of the full length ORF1ab ( 21k in length), ORF1a and ORF1b were defined by PROKKA as two separate genes. Notably, ORF1a, ORF3a, ORF6, and ORF8 and S, show strong separations between nodes from different species. ORF8 from 3 bat-CoV co-cluster with ORF8 from SARS-CoV-2 (RaTG13, bat-SL-CoVZC45 and bat-SL-CoVZXC21 respectively). The remaining bat-CoV ORF8 do not co-cluster with SARS-CoV-2 ORF8 even without the edge filtering threshold. For S, the bat-CoV RaTG13 co-cluster with COVID-19 and pangolin. A cluster of bat-CoVs break off for ORF1b and M, suggesting a large amount of variation amongst bat-CoV for these genes.
Interestingly, at the whole-genome level, SARS-CoV-1 and MERS-CoV only share 79.5% and 50.0% sequence similarity with SARS-CoV-2. On the other hand, SARS-CoV-2-like coronaviruses found in pangolins (pangolin-CoVs) and the bat-CoV RaTG13 share 91.0% and 96.0% similarity, respectively.
The potential role of bats and pangolins as reservoir species in the emergence of SARS-CoV-2, as well as the role other intermediary hosts potentially played, has spurred a number of research approaches and collaborations between experts of different fields.
As such, the current study was carried out as part of a “CoronaHack” hackathon event that took place in April 2020. There, the authors gained access to all the genomes and related metadata that was available at the time (between December 2019 and April 2020).
What did the researchers do?
The team employed a number of contemporary methodologies to analyze a wide range of genomic sequences isolated from human SARS-CoV-2 (n=163), bats (n=215), and pangolins (n=7).
The sequences were systematically compared at the whole-genome, gene, codon usage and variant levels to investigate the similarities and differences that exist across 89 different host species.
What did they find?
At the whole-genome levels, bat-CoV RaTG13 still shared the most similarity with SARS-CoV-2. However, all 7 pangolin-CoV genomes were more closely related to SARS-CoV-2 than the remaining 214 bat-CoV genomes.
“This relationship has previously been reported, and a recombination event between pangolin-CoVs and RaTG13 has been theorized,” say Shih and colleagues.
Gene-gene network analysis showed strong host-associated divergences in ORF3a, ORF6, ORF7a, ORF8 and the spike (S) protein. Strong host-species separations were also observed in codon usage bias profiles.
For example, three bat-CoV ORF8 genes were more similar to SARS-CoV-2 than most of the pangolin-CoV ORF8 genes.
By contrast, the S genes of pangolin-CoV and SARS-CoV-2 were more similar to each other (97.5%), than the S genes of RaTG13 and SARS-CoV-2 (95.4%).
“This is significant as the S protein plays an important role in the initial penetration and infection of host cell,” say the researchers.
However, the S gene in RaTG13 was still more similar to that of SARS-CoV-2 than to those of all other bat-CoVs analyzed in this study, they add
“This supports the theory that neither a currently sequenced pangolin-CoV or bat-CoV is the most recent ancestor of SARS-CoV-2,” writes the team.
The researchers identified strong host-species separation in the overall codon usage when multiple genes were combined in the analysis.
They found very little variation in codon usage bias within the SARS-CoV-2 isolates, but all pangolin-CoVs and three bat-CoVs had more similar codon usage to SARS-CoV-2.
Identifying high-impact variants
The team also identified several high-impact variants in bat-CoV samples, including a stop-gain for ORF10 and inframe insertions and deletions for the nucleocapsid (N) protein.
Importantly, the stop-gain was identified at amino acid position 26 in ORF10 among 57 of the 59 bat-CoV genomes, where ORF10 shared more than 80% similarity with SARS-CoV-2.
In a previous study of SAR-CoV-2 and pangolin CoV genomes, position 26 was also identified as a region of population-level variation, say Shih and colleagues.
In the N gene, the team observed multiple inframe variants for the same amino acid position in two groups of bat-CoVs. The analysis revealed two inframe insertions at amino acid position 7 and two inframe deletions at positions 238 and 385.
What are the study implications?
“These naturally occurring variants we observed across bat-CoV and pangolin-CoV may be associated with selection advantages, such as virulence or the efficiency infect a specific host species,” suggest Shih and colleagues.
The researchers say the study has revealed a high degree of host-species separation in ORF3a, ORF6, ORF7a, ORF8 and S, as well as in codon usage.
It has also identified a number of amino acid positions that demonstrate high impact variants in several bat-CoVs and pangolin-CoVs.
“These are potentially functionally important positions of the protein and warrant further research,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources