We are now well aware that minks and ferrets can be easily infected with SARS-CoV-2 and that keeping farmed minks in a high-density environment may prompt SARS-CoV-2 proliferation. This may consequently select viral mutants with the propensity to jeopardize the efficacy of potential drugs and vaccine candidates against COVID-19.
Such natural selection "adaptation" in SARS-CoV-2 may arise during coronavirus amplification in farmed minks by introducing mutations not specific for the viral cycle in humans. Furthermore, the infection with a specific mutant strain of SARS-CoV-2 from farmed minks – known as Y453F – can be widely spread among humans.
More specifically, this strain harbors amino acid mutation Y453F in the sequence that encodes spike glycoprotein and has been found in approximately 300 viral sequences isolated from humans in the Netherlands and around Europe, but also in minks.
This is why a research group led by Dr. Takuma Hayashi from the National Hospital Organization Kyoto Medical Center and Japan Science and Technology Agency in Tokyo, decided to investigate the virologic characteristics of the aforementioned SARS-CoV-2 mutant with the use of three-dimensional protein structural analysis.
The use of complex modeling programs
A vast amount of data regarding the three-dimensional structure of the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein was used in their research approach. This was coupled with the data on the three-dimensional structure of six neutralizing antibodies known to bind to the spike glycoprotein of the virus.
The structure of the mutant was predicted with the use of the Spanner program. In short, Spanner utilizes structural fragments in order to generate a hybrid template by querying a database of structural fragments (primarily relying on the geometry of the endpoints of the gap).
Finally, the researchers have evaluated binding characteristics of the spike glycoprotein Y453F viral mutant to human angiotensin-converting enzyme 2 (ACE2) and appraised its affinity to six neutralizing monoclonal antibodies employing the MOE modeling program and Cn3D macromolecular structure viewer.
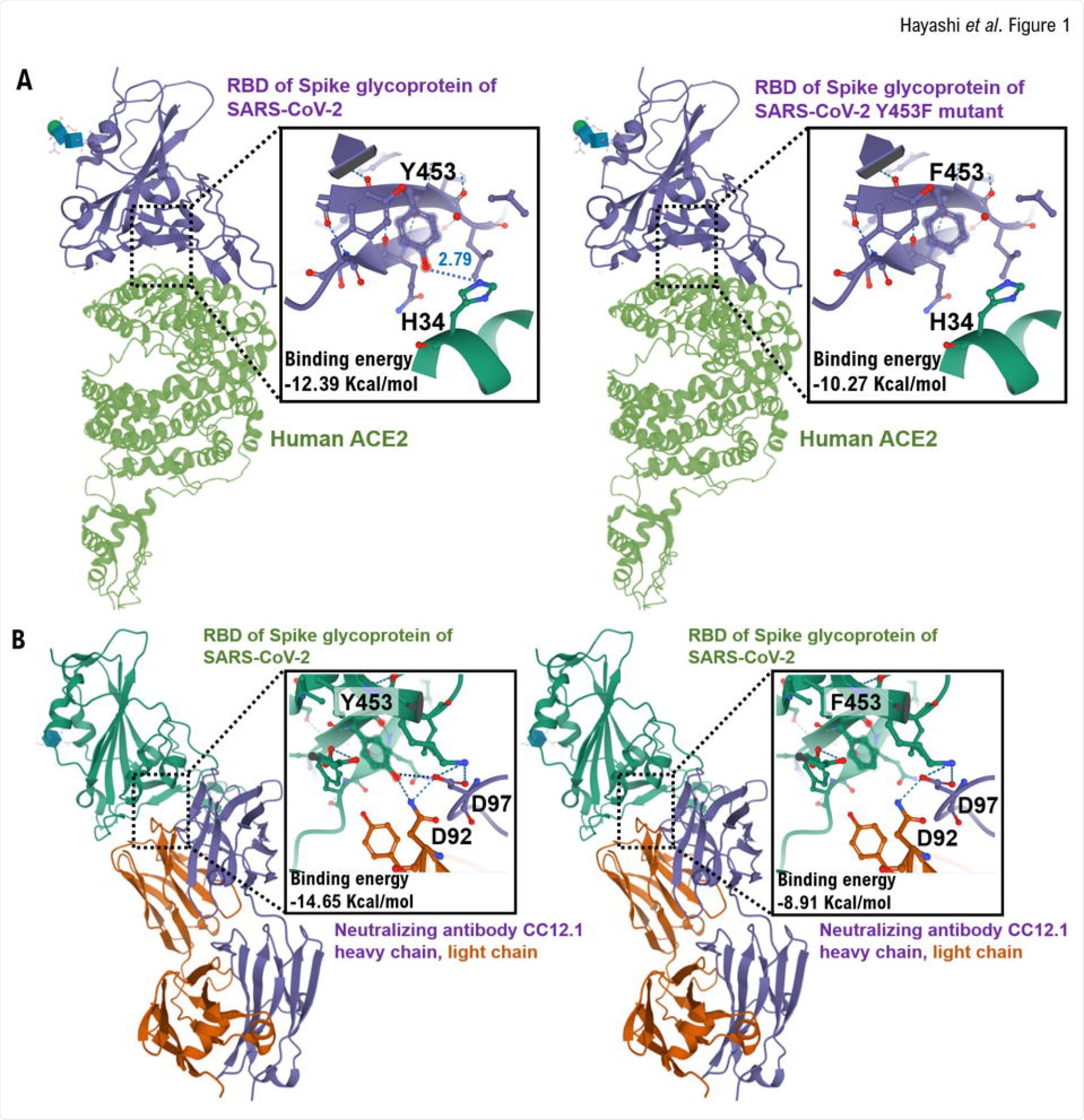
Interactions between the RBD and RBD Y453F mutant in the spike glycoprotein of SARS-CoV-2 and heavy chain of neutralizing monoclonal antibody (CC12.2). (A) The interaction between Angiotensin-converting enzyme 2 (ACE2) (green) and residues of the conventional receptor-binding domain (RBD) or RBD Y453F mutant (purple) is shown using the three-dimensional structure model. It is speculated that the Y453 amino acid residue of the conventional RBD is hydrogen-bonded to the H34 amino acid residue of human ACE2. However, the binding between the F453 amino acid residue of the RBD mutant and the H34 amino acid residue of human ACE2 is presumed to be slightly weak. From these results, the affinity between the spike glycoprotein of the RBD Y453F mutant and human ACE2 is presumed to be slightly weak compared with the conventional RBD. (B) The interaction between the heavy chain (purple) and light chain (brown) of neutralizing monoclonal antibody CC12.1 and residues of the conventional RBD or RBD Y453F mutant (green) is shown using the three-dimensional structure model. It is speculated that the Y453 amino acid residue of the conventional RBD is hydrogen-bonded to the D92 amino acid residue of the light chain and D97 amino acid residue of the heavy chain of neutralizing monoclonal antibody CC12.1. However, the binding between the F453 amino acid residue of the RBD mutant and the D92 and D97 amino acid residues of neutralizing monoclonal antibody CC12.1 is presumed to be weak. From these results, the affinity between the spike glycoprotein of the RBD Y453F mutant and neutralizing monoclonal antibody CC12.1 is presumed to be low compared with the conventional RBD. The three-dimensional structure models are shown by Cn3D macromolecular structure viewer.
Escaping antibody detection
In short, the Y453F mutation did not have an impact on the three-dimensional structure of conventional SARS-CoV-2 spike glycoproteins. Moreover, the study has shown that the spike glycoprotein Y453F mutant binding to human ACE2 was somewhat weaker in comparison to the conventional SARS-CoV-2 spike glycoprotein.
Also, this study demonstrated that the affinity between the spike glycoprotein Y453F mutant and four of the six tested monoclonal antibodies was plainly weak when compared to the conventional SARS-CoV-2 spike glycoprotein, suggesting that this mutation may be used to escape detection from neutralizing antibodies.
Therefore, it is thought that the affinity between corresponding amino acid residues in the variable region of the antibody and the spike glycoprotein of the Y453F SARS-CoV-2 mutant decreased due to inadequate recognition of the monoclonal antibody to spike glycoproteins.
Minks as a potential cause of new infectious wave
Taking these results into account, we have to bear in mind that data on all SARS-CoV-2 mutants have thus far not been published. Hence, it is still a puzzle whether SARS-CoV-2 mutants in people working on mink farms actually stem from the farmed minks.
Nevertheless, in this study, the subspecies of SARS-CoV2 that has been derived from farmed minks were actually detected in the group of infected individuals, strengthening the hypothesis implicating this transmission pathway.
"Mutations in SARS-CoV-2 that lead to generation of SARS-CoV-2 subspecies have made humans and animals susceptible to infection through easy propagation in the host, thereby making it difficult to identify the effects of therapeutic agents or vaccines for COVID-19", accentuate study authors in this bioRxiv paper.
In any case, SARS-CoV-2 variants in millions of infected farmed mink are basically uncontrolled, which is a valid reason for concern that infection with SARS-CoV-2 mutants may cause serious symptoms in humans and elicit another wave of the COVID-19 pandemic if we are not careful and vigilant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources