The function of proteins is tightly dependent on their structure and is highly sensitive to an ambient environment. It is crucial to have a complete biophysical characterization of proteins, specifically in drug-hunting endeavors.
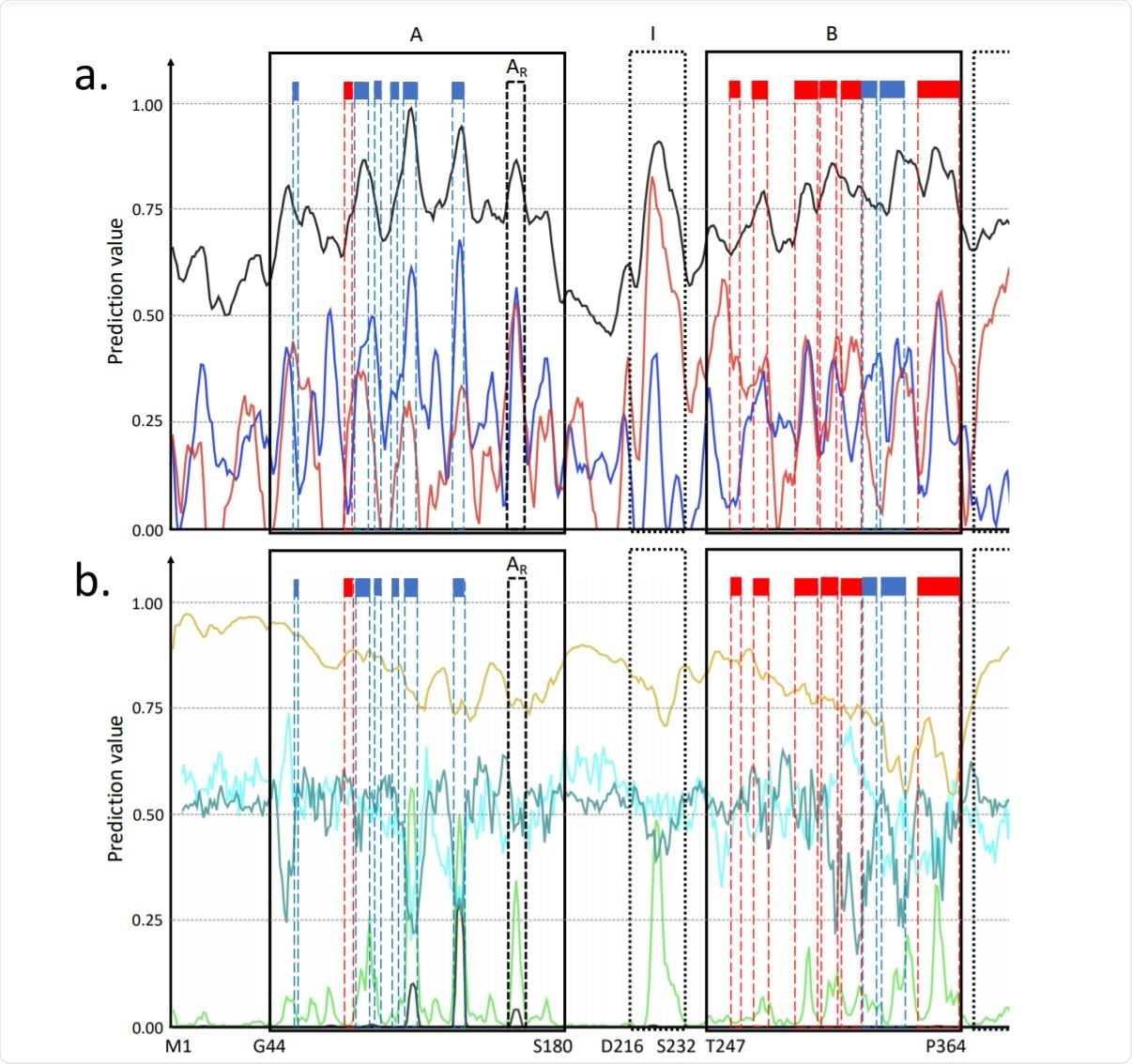
Predictions for the P0DTC9 SARS-CoV-2 protein amino acids. Image Credit: bioRxiv

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19), consists of an assembly of proteins that determine its infectious and immunological behavior. These proteins determine their response to therapeutics.
Not all the SARS-CoV-2 proteins or regions have a well-defined three-dimensional structure. Many proteins exhibit ambiguous, dynamic behavior that is not evident from static structure representations generated by structural biology approaches or molecular dynamics simulations using these structures.
To identify behavior or features of these proteins that might not be captured by structural biology or molecular dynamics approaches, Luciano Kagami et al. provide protein-sequence-based predictions of the backbone and side-chain dynamics and conformational propensities of these proteins, as well as derived early folding, disorder, b-sheet aggregation, and protein-protein interaction propensities. They present a website (http://sars2.bio2byte.be/) that provides this information for researchers.
The website is approved as an ELIXIR-Belgium emerging service in 2020. The website's information was visualized online using the Django framework, with the ApexCharts JavaScript library employed for visualization of the predictions and their MSA distribution. This work has received funding from the European Union’s Horizon 2020 research and the innovation program under the Marie Skłodowska-Curie grant. It was recently published in the bioRxiv* preprint server.
They targeted amino acid sequences of the 14 proteins, obtaining the multiple sequence alignments (MSAs) for these sequences using a BLAST search from UniProt and applying default parameters against the Uniref90 protein dataset. They followed this by the standard UniProt ClustalW alignment procedure to obtain the MSA.
The authors predict the backbone dynamics (DynaMine) and related side-chain dynamics and conformational propensities at the individual amino acid level. The study included early folding (EFoldMine), disorder (DisoMine), beta-sheet aggregation (Agmata), protein-protein interactions (SeRenDIP), and SeRenDIP-CE conformational epitope propensities. A detailed description of each prediction per-protein is available on their website.
In this study, the predictions attempt to capture the 'emergent' properties of the proteins based on the inherent biophysical propensities encoded in the sequence. This approach has its advantages as opposed to the context-dependent behavior (such as the final folded state). For example, the authors show how they detect remote SARS-CoV-2 protein homologs by biophysical similarity, giving more accurate results than directly using amino acid information.
The authors show the biophysical variations observed in homologous SARS-CoV-2 proteins. The study indicates the likely limits of the functionally relevant biophysical behavior of the proteins.
Luciano Kagami et al. presents predictions for the P0DTC9 protein - a nucleoprotein of 419 amino acids with both monomeric and oligomeric forms that interact with RNA and protein M and NSP3. These interactions are essential during the early stage of infection.
A detailed description of the wide propensities for this protein is given. The authors also discuss the predictions for a region where there is no structural or functional information available. It is important to note that this study provides the nitty-gritty around the biophysical predictions for a protein under investigation, which may be used for further diverse applications.
Therefore, the authors provide researchers with information on their website on the possible behaviors of SARS-CoV-2 proteins that are not evident from the static models generated by structural biology nor from molecular dynamics simulations based on models.
These predictions reflect ‘emerging’ properties based on the sequence. A different perspective exploring the SARS-CoV-2 proteins is at the disposal of researchers. This study should help us further to understand the mode of action of the overall virus, the authors write.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Online biophysical predictions for SARS-CoV-2 proteins; Luciano Kagami, Joel Roca-Martínez, Jose Gavaldá-García, Pathmanaban Ramasamy, K. Anton Feenstra, Wim Vranken bioRxiv 2020.12.04.411744; doi: https://doi.org/10.1101/2020.12.04.411744
- Peer reviewed and published scientific report.
Kagami, Luciano, Joel Roca-Martínez, Jose Gavaldá-García, Pathmanaban Ramasamy, K. Anton Feenstra, and Wim F. Vranken. 2021. “Online Biophysical Predictions for SARS-CoV-2 Proteins.” BMC Molecular and Cell Biology 22 (1). https://doi.org/10.1186/s12860-021-00362-w. https://bmcmolcellbiol.biomedcentral.com/articles/10.1186/s12860-021-00362-w.