There is much interest surrounding the nature and determinants of immunity following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. It is known that adaptive specific immunity is rapidly elicited on exposure to the virus, involving both T cells and antibody-producing B cells.
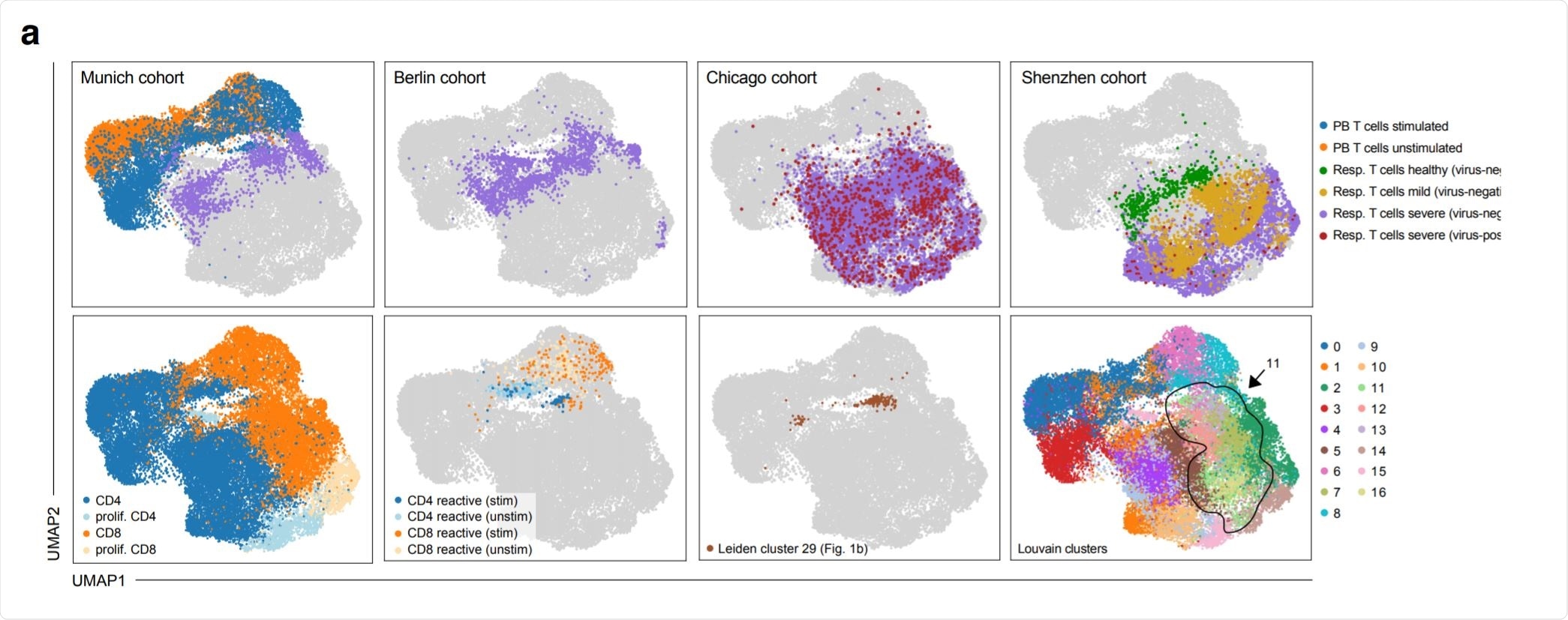
Phenotypic convergence of in vitro stimulated reactive T cells from peripheral blood and in vivo T cells from the respiratory tract of severely diseased patients. Image Credit: https://www.medrxiv.org/content/10.1101/2020.12.07.20245274v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Issues with T Cell Phenotyping Attempts
The unprecedented speed of research has led to the identification of immunodominant SARS-CoV-2 antigens and the corresponding antigen-reactive T cells. As of now, the methods used to specify the extent of activation and differentiation of the T cells are varied, leading to confusion about many phenotypic aspects. Some have, for instance, reported the phenotypes of activated T cells alone, without a baseline characterization of inactivated T cells in the same situation. In other studies, the specificity of the antigens associated with the activation profiles characterized by the researchers remains unverified.
The challenges of phenotyping only activated T cells that have reacted to specific antigens include the fact that these antigen-reactive cells have been restimulated with SARS-CoV-2 antigens in vitro. Only so can such cells be detected. This is followed by identifying upregulated surface markers of activation, such as CD154, or activation cytokines such as the well-known activation marker IFNγ.
However, the process of restimulation automatically entails major shifts in the phenotype. This is because in vitro activation of T cells is associated with a distinctive expression profile compared to unperturbed T cells, here termed in vivo T cells.
pHLA Multimers – and Their Limitations
One of the tools used here to prevent such unhelpful differences is the use of specifically engineered peptide human leukocyte antigen (pHLA) multimers, that can be removed to leave unperturbed antigen-reactive T cells for phenotyping.
These multimers can only be developed if the specific epitope is properly defined first, and each fits only one HLA haplotype. HLA class II CD4 T cells are difficult to purify with this technology, which presents a hurdle to the phenotypic profiling of antigen-reactive T cells without significant changes due to restimulation in vitro.
Indiscriminate T Cell Activation Data
Many researchers have used data from the immunophenotypic gene atlas found in the Human Cell Atlas Initiative. However, this contains a host of different phenotypes from different disease states and various tissue sites.
Again, antigen-reactive T cells will have different activation states depending on their location in the body, as all are not exposed to the infectious agent. Even in COVID-19, T cells in the lung or upper airway might be expected to be more activate compared to those in the circulation, since the latter are not exposed to viral antigens.
Single Cell RNA Sequencing
What is required for in vivo activated T cell phenotyping is clinical data and deep profiling of T cells exposed to specific antigens, across different tissues, at the same time, from the same patient.
For this purpose, the current researchers used single-cell RNA sequencing to examine the transcriptional patterns of T cells all over the body, as well as to define T cell receptor (TCR) sequences, at the same time. This technique is capable of generating phenotypes of antigen-reactive T cells in a state of activation.
Reverse Phenotyping
The investigators first isolated T cells from different tissues, and for each sample, they split them up into different groups. Each group represents the T cells that react to a common specific antibody and have the same unique TCR. Some of these groups were stimulated in vitro by specific antigens, while others were left as such, to serve as comparisons.
Both groups were then sequenced, with the unstimulated TCR being the reference standard to identify the T cells belonging to the same group, stimulated or not. This practice is referred to as reverse phenotyping, as the changes in phenotype are used to identify the effects of a genetic change.
This method was followed by the researchers to identify and study T cells reactive to SARs-CoV-2 spike protein peptides. The cells came from the peripheral blood of severe COVID-19 patients. Some of the cells were stimulated by the peptide mix, while antigen-presenting cells from the same patient ensured proper antigen processing and presentation for T cell activation.
RNA sequencing showed specific transcriptional patterns in the T cell populations responding to each of the antigens in the mixture. They carried out phenotyping of CD8 and many T helper CD4 cells at the same time, thus ensuring the testing was haplotype- and epitope-independent. Simultaneous testing of many different TCRs was conducted for antigen reactivity and functional profiling.
Next, they exploited CRISPR/Cas9 technology to produce T cells expressing transgenic TCRs, enabling them to validate the spike antigen-TCR reactivity of the TCRs from specific clonotypes.
Restimulation May Produce Confounding Changes
Upon stimulation, CD4 T cells have been described to show an early activation and late activation response, characterized by an IFN response and T cell proliferation, respectively.
In the present analysis, a single cluster of CD4 cells showing a high proliferation index was observed, but many other cells showed a seemingly nonspecific transcriptional shift. However, the latter were those with an early activation profile.
CD8 T cells also show an early and late activation profile, namely, high cytotoxicity and cytokine secretion phases. In this study, the first profile was seen with almost all CD8 T cells, but only one cluster showed the second profile. This cluster showed upregulated genes such as TNF and IFNγ. They observed that all reactive T cells, whether CD4 or CD8, converged on this cluster.
In response to stimulation, therefore, some activation markers are increased, as expected, and others show more complex swings in marker expression outside supposedly stable phenotypes. Restimulated CD4 T cells expressed effector cytokines like TNF and IFNγ, as well as CD137 marker.
They also expressed co-inhibitory molecules such as PD1 and LAG3, which were earlier described to be characteristic of T cell exhaustion in severe disease. Instead, the current study indicates that these are markers of T cell activation.
Many induced markers show differential transcription shifts across the board, within clonotypes, and also between stimulated vs unstimulated and reactive vs non-reactive clonotypes overall.
This supports the confounding influence of in vitro restimulation on T cell phenotypes.
What are the Implications?
The importance of this study lies first in its demonstration of the systematic shifts in phenotype as a result of antigen stimulation, by comparing the transcriptional profile in antigen-reactive and antigen-unreactive clonotypes, both before and after stimulation. The researchers were able to define the unperturbed in vivo phenotypes of T cells activated by SARS-CoV-2 antigens.
Thirdly, they compared T cells from patients with COVID-19, taken from various sources in the respiratory tract, from patients with varying disease severities, and at different time points, with those taken from healthy controls. They found that respiratory T cells from COVID-19 patients were most similar, in their transcriptional pattern, to T cells from peripheral blood after being exposed to SARS-CoV-2 antigens in vitro. This indicates they say, “a 'hot' phenotype driven by acute activation at the site of infection.”
On the other hand, the transcriptional phenotype of respiratory T cells from virus-negative severe COVID-19 patients matched those of unperturbed antigen-reactive T cells from the peripheral blood, indicating their ‘cold’ phenotype.
We present transcriptional shifts from acute disease to resolution after virus clearance in antigen reactive CD4 and CD8 T cells during the course of COVID-19.”
Using reverse phenotyping, they were able to recreate the antigen-reactive clonotypes as they would have been without restimulation. This enabled them to “investigate systematic phenotypic effects that are introduced by antigenic re-stimulation in vitro, and on the other hand, explore the unperturbed in vivo phenotype of antigen-reactive T cells.”
This tool could help understand adaptive immunity following viral infection, helping to develop better therapies and vaccines for newly emerging pathogens, as well as SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Fischer, D. S. et al. (2020). Single-cell RNA sequencing reveals in vivo signatures of SARS-CoV-2-reactive T cells through 'reverse phenotyping'. medRxiv preprint. doi: https://doi.org/10.1101/2020.12.07.20245274. https://www.medrxiv.org/content/10.1101/2020.12.07.20245274v1
- Peer reviewed and published scientific report.
Fischer, David S., Meshal Ansari, Karolin I. Wagner, Sebastian Jarosch, Yiqi Huang, Christoph H. Mayr, Maximilian Strunz, et al. 2021. “Single-Cell RNA Sequencing Reveals Ex Vivo Signatures of SARS-CoV-2-Reactive T Cells through ‘Reverse Phenotyping.’” Nature Communications 12 (1): 4515. https://doi.org/10.1038/s41467-021-24730-4. https://www.nature.com/articles/s41467-021-24730-4.