There is a huge and growing demand for better and more cost-effective molecular diagnostic tests for COVID-19, as the pandemic shows few signs of coming under control in the near future.
The most commonly used test is the reverse transcriptase-polymerase chain reaction (RT PCR), which uses one or multiple gene targets. A new study published in the preprint medRxiv* server in December 2020 discusses the utility of using different primer sets targeting different regions of the N gene target.
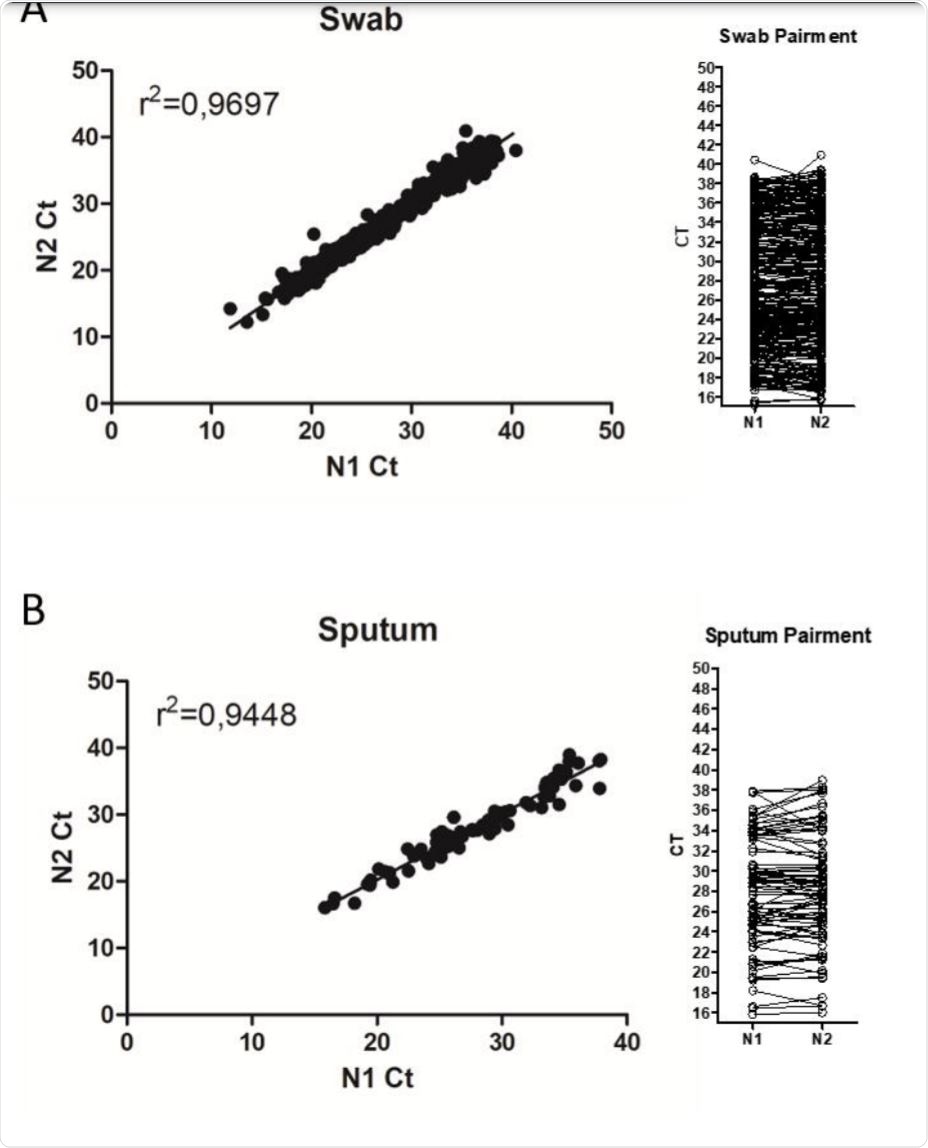
Comparison between N1 and N2 Ct values during SARS-CoV-2 detection in patient samples. Image Credit: https://www.medrxiv.org/content/10.1101/2020.12.06.20244905v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
CDC Kit Uses Double Probe-Primer Set
Earlier studies have shown different results from the use of various probes and primers. Diagnostic kits from the US Centers for Disease Control and Prevention (CDC) target the N antigen and use two primer sets that have high sensitivity for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Several tests use only a single set of primers for SARS-CoV-2 detection, but there is no real evidence for or against this practice, in contrast to the use of targeting different regions of the virus genome.
The justification for using multiple targets is the high frequency of alteration in the viral genome that can make the primer less capable of viral detection. However, this practice pushed up the level of technology and the cost of the test. The researchers failed to detect any mutation in the target regions of the N gene, in genomes collected from South America.
The current paper describes the results using either of the two N commercial antigens in the CDC test panel, which is considered to be among the most sensitive and specific in the world.
Study Details
The researchers used over 1,000 samples of nasopharyngeal, nasal, and oropharyngeal swabs and sputum, collected in May and June 2020, all residual samples used for SARS-CoV-2 detection in the Laboratory of Vectors and Infection Disease. They found that the cycle threshold (Ct) values were similar for all samples with either N1 or N2 primer sets.
They assessed the frequencies of results between N1 and N2 primer sets quantitatively, to arrive at the fitness of each range of Ct values. They found that either set was capable of detecting viral RNA with the same Ct value. This agrees with earlier studies on the performance of primer-probe sets from several diagnostic kits.
This suggests that a single primer set could be used to test patients with this infection. This could reduce testing costs while maintaining diagnostic accuracy and efficiency. Some studies have even concluded that saliva could be used for diagnosis without loss of sensitivity for viral detection in COVID-19.
Target Accuracy
The researchers also attempted to determine whether the Ct value cut-offs were appropriate. Using commercially available N gene templates, they measured the limits of detection (LoD) and cut-offs for each primer set. They found that either was capable of detecting 5 genome copies per microliter of reaction. The Ct value was ~34 for either N1 or N2.
Many modeling studies suggest the CDC test uses a high Ct cut off, with standard protocols that recommend a Ct of 40. The current study shows that either primer gives rise to nonspecific amplification, perhaps due to the annealing of primers in different regions of the human genome.
The researchers suggest the use of the conventional PCR technique with acrylamide gel in remote locations where the molecular diagnosis may be difficult to access.
The researchers also evaluated samples from a single patient, collected 2-6 days after the first. Only about a third were negative, another third had a reduced Ct value, and the rest were positive at the same Ct value. For doubly positive samples with a Ct >33, the test had low sensitivity and specificity.
Thus, samples that are tested with a Ct that is near the LoD are poorly predictive of a true positive result. Instead of using these samples for diagnosis, new samples should be obtained for a second confirmatory test.
Poor Predictive Value at Higher Ct
A prior study showed that the Ct value increased over the 21-day interval between successive tests. Values higher than 40 are sometimes recommended to be discarded in a diagnostic test for this coronavirus. Both the current study and other earlier studies show that the predictive value of the RT PCR test falls significantly when the Ct is above 35, using the CDC protocol.
Patients in the current study whose samples tested positive with a Ct above 33 had very different test results when the tests were repeated in 2-6 days. Thus, at high Ct values, positive tests may have a low predictive value. In other words, samples that test positive at high Ct values are poorly reproducible.
The reasons for this could be cross-contamination of samples, low viral load due to the late or very early phase of infection, and low viral load near the LoD of the testing method in use. In such cases, a new sample should be obtained and tested for a reliable diagnosis.
Insights into True vs False Reinfection
Several cases of reinfection have been reported, in some of which the person first had a case of mild SARS-CoV-2 infection with a low viral load and a test positive with a high Ct value. This is followed by a period in which seroconversion does not occur, after which a second infection is reported, this time with a high viral load and with severe symptoms. At this point, the patient seroconverts.
One of the best studies on humoral immunity in SARS-CoV-2 infection has shown that neutralizing antibodies are produced robustly and durably after a single infection. Given these findings, the authors of the current paper suggests that if high Ct values are reported at the time of the initial or second diagnosis in such cases, a verdict of reinfection should be withheld.
To uphold this diagnosis of reinfection, both episodes should have a history of a clear viral load and no evidence of cross-contamination of samples during processing, which would account for both the high Ct values and the differences in the target genes. Only if the viral load in both episodes corresponds to a Ct value below 30 should it be considered to be a case of reinfection, they say.
What are the Conclusions?
The current study thus shows that either N1 or N2 sets of primers and probes can be used individually to diagnose this infection. However, with the CDC testing kit, the sensitivity and specificity drop drastically when the Ct values are close to the LoD of each laboratory. Thus, the CDC RT PCR protocol should be used with care in such cases.
Instead, in such samples, if the Ct is above the LoD, the RNA should be re-extracted and another protocol used. If there is any doubt, a fresh sample should be collected before arriving at a positive diagnosis. Thus, this study could affect the interpretation of future test results in COVID-19 as well as the methods of sample analysis by PCR targeting the N gene.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Klein, R. C. et al. (2020). Optimizing SARS-CoV-2 molecular diagnostic using N gene target: insights about reinfection. medRxiv preprint. doi: https://doi.org/10.1101/2020.12.06.20244905.
https://www.medrxiv.org/content/10.1101/2020.12.06.20244905v1
- Peer reviewed and published scientific report.
Klein, Raphael Contelli, Mary Hellen Fabres Klein, Larissa Gomes Barbosa, Lívia Vasconcelos Gonzaga Knnup, Larissa Paola Rodrigues Venâncio, Jonilson Berlink Lima, and Théo Araújo-Santos. 2021. “Identifying Inconclusive Data in the SARS-CoV-2 Molecular Diagnostic Using Nucleocapsid Phosphoprotein Gene as a Target.” Archives of Pathology & Laboratory Medicine 146 (3): 272–77. https://doi.org/10.5858/arpa.2021-0423-sa. https://meridian.allenpress.com/aplm/article/146/3/272/473998/Identifying-Inconclusive-Data-in-the-SARS-CoV-2.